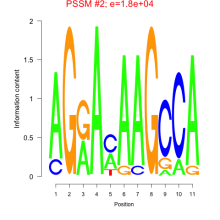
Phatr_bicluster_0088 Residual: 0.33
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0088 | 0.33 | Phaeodactylum tricornutum |
Displaying 1 - 37 of 37
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_13233 diguanylate cyclase with gaf sensor
PHATRDRAFT_14908 rna polymerase sigma factor (Sig70-cyanoRpoD)
PHATRDRAFT_16540 aldehyde dehydrogenase 3member b1 (ALDH_F3-13-14_CALDH-like)
PHATRDRAFT_19518 serine racemase (PRK08198)
PHATRDRAFT_22315 mitochondrial carrier protein (Mito_carr)
PHATRDRAFT_31436 peroxisomal membrane protein22kda (Mpv17_PMP22)
PHATRDRAFT_31925 glutathione s-transferase omegaisoform cra_a (GST_C_family)
PHATRDRAFT_33920 (MerC)
PHATRDRAFT_34957 selw selh selenoprotein (Rdx)
PHATRDRAFT_37532 (SRPBCC superfamily)
PHATRDRAFT_38534 at3g06050 f24f17_3 (Thioredoxin_like superfamily)
PHATRDRAFT_38634 retinol binding proteincellular
PHATRDRAFT_40695 cog0596: hydrolases or acyltransferases (alpha beta hydrolase superfamily) (Abhydrolase_6)
PHATRDRAFT_43896 atp-dependent clp protease adaptor protein (ClpS)
PHATRDRAFT_44704 protein n-terminal asparagine amidohydrolase (N_Asn_amidohyd superfamily)
PHATRDRAFT_44968 ectonucleoside triphosphate diphosphohydrolase 4 (GDA1_CD39 superfamily)
PHATRDRAFT_46768 epsk domain protein (Branch superfamily)
PHATRDRAFT_47188 nod3isoform cra_d (LRR_RI superfamily)
PHATRDRAFT_47439 f-box only protein 31 (DUF3506 superfamily)
PHATRDRAFT_48225 nod2 protein (LRR_RI superfamily)
PHATRDRAFT_49097 vitelliform macular dystrophy 2-like protein 2 (Bestrophin superfamily)
PHATRDRAFT_49336 gcn5-related n-acetyltransferase (NAT_SF)
PHATRDRAFT_50481 heat shock factor rhsf2 (HSF_DNA-bind superfamily)
PHATRDRAFT_8167 thioredoxin (TRX_family)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006352 | transcription initiation | 0.0205774 | 1 | Phatr_bicluster_0088 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0510752 | 1 | Phatr_bicluster_0088 |
| GO:0008152 | metabolism | 0.0865285 | 1 | Phatr_bicluster_0088 |
| GO:0006810 | transport | 0.287164 | 1 | Phatr_bicluster_0088 |
| GO:0006118 | electron transport | 0.33492 | 1 | Phatr_bicluster_0088 |


Comments