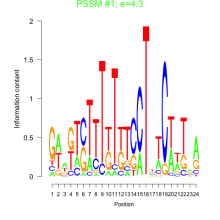
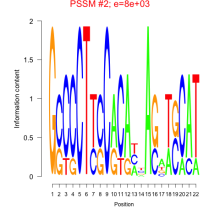
Phatr_bicluster_0089 Residual: 0.28
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0089 | 0.28 | Phaeodactylum tricornutum |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_13047 (CAP_C)
PHATRDRAFT_18067 human ran binding protein 16-like (IBN_N)
PHATRDRAFT_19901 cg8363-isoform a (nt_trans superfamily)
PHATRDRAFT_34457 mgc80931 protein
PHATRDRAFT_36060 methyltransferase protein (Methyltransf_21)
PHATRDRAFT_42458 phosphoserine aminotransferase (AAT_I)
PHATRDRAFT_45120 myo-inositol-1-phosphate synthase (PLN02438)
PHATRDRAFT_45476 actin binding protein (VHP superfamily)
PHATRDRAFT_45580 coronin 6 clipin e type c isoform 1 (DUF1900)
PHATRDRAFT_46109 core alpha-fucosyltransferase (Glyco_transf_10 superfamily)
PHATRDRAFT_47010 transducin family protein wd-40 repeat family protein (WD40 superfamily)
PHATRDRAFT_47314 (Spc97_Spc98)
PHATRDRAFT_48381 actin cortical patch (WD40 superfamily)
PHATRDRAFT_48491 alkaline serine protease (Peptidases_S8_PCSK9_ProteinaseK_like)
PHATRDRAFT_48622 aspartyl asparaginyl beta-hydroxylase
PHATRDRAFT_48795 silk fibroin
PHATRDRAFT_50095 exostosin family protein (Exostosin)
PHATRDRAFT_50463 16isoform cra_c (Peptidase_S28 superfamily)
PHATRDRAFT_54169 glucosidase ii beta subunit (PRKCSH superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007010 | cytoskeleton organization and biogenesis | 0.00246975 | 1 | Phatr_bicluster_0089 |
| GO:0006021 | myo-inositol biosynthesis | 0.00246975 | 1 | Phatr_bicluster_0089 |
| GO:0000226 | microtubule cytoskeleton organization and biogenesis | 0.004934 | 1 | Phatr_bicluster_0089 |
| GO:0000103 | sulfate assimilation | 0.004934 | 1 | Phatr_bicluster_0089 |
| GO:0000059 | protein-nucleus import, docking | 0.0122939 | 1 | Phatr_bicluster_0089 |
| GO:0008654 | phospholipid biosynthesis | 0.0220309 | 1 | Phatr_bicluster_0089 |
| GO:0006508 | proteolysis and peptidolysis | 0.1023 | 1 | Phatr_bicluster_0089 |
| GO:0009058 | biosynthesis | 0.105854 | 1 | Phatr_bicluster_0089 |
| GO:0008152 | metabolism | 0.529843 | 1 | Phatr_bicluster_0089 |

Comments