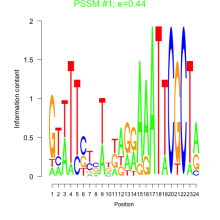
Phatr_bicluster_0129 Residual: 0.31
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0129 | 0.31 | Phaeodactylum tricornutum |
Displaying 1 - 28 of 28
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10925 ubiquitin-like protein 5 (UBQ superfamily)
PHATRDRAFT_16067 protein kinase domain containing protein (S_TKc)
PHATRDRAFT_16509 complex 1lyr family (Complex1_LYR_1)
PHATRDRAFT_21970 biopterin transport-related protein bt1 (BT1)
PHATRDRAFT_23988 cytosolic glucose-6-phosphate isomerase (PLN02649)
PHATRDRAFT_29551 (MPP_superfamily superfamily)
PHATRDRAFT_36358 (NYN_YacP superfamily)
PHATRDRAFT_36534 dtw domain (DTW superfamily)
PHATRDRAFT_40369 pdz domain protein (PDZ_signaling)
PHATRDRAFT_40489 (Smr)
PHATRDRAFT_43163 pentatricopeptiderepeat-containing protein
PHATRDRAFT_43907 chromosome 20 open reading frame 67 (PCIF1_WW superfamily)
PHATRDRAFT_46348 choline o-acetyltransferase(choline acetylase) (ANK)
PHATRDRAFT_46398 (2OG-FeII_Oxy_5 superfamily)
PHATRDRAFT_48450 ring zinc finger protein (RING)
PHATRDRAFT_50618 (DUF1279 superfamily)
PHATRDRAFT_54088 cell surface flocculin
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006094 | gluconeogenesis | 0.00861852 | 1 | Phatr_bicluster_0129 |
| GO:0006298 | mismatch repair | 0.0103346 | 1 | Phatr_bicluster_0129 |
| GO:0006096 | glycolysis | 0.0590046 | 1 | Phatr_bicluster_0129 |
| GO:0006464 | protein modification | 0.104035 | 1 | Phatr_bicluster_0129 |
| GO:0016567 | protein ubiquitination | 0.118111 | 1 | Phatr_bicluster_0129 |
| GO:0006468 | protein amino acid phosphorylation | 0.262204 | 1 | Phatr_bicluster_0129 |
| GO:0006118 | electron transport | 0.33492 | 1 | Phatr_bicluster_0129 |


Comments