Phatr_bicluster_0156 Residual: 0.31
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0156 | 0.31 | Phaeodactylum tricornutum |
Displaying 1 - 26 of 26
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10089 hal3a protein (Flavoprotein superfamily)
PHATRDRAFT_10102 (RRM_CNOT4)
PHATRDRAFT_11352 membrance occupation and recognition nexus protein 1 (MORN superfamily)
PHATRDRAFT_12595 methionyl aminopeptidase 1 (PLN03158)
PHATRDRAFT_12868 eukaryotic translation initiation factor 1b (eIF1_SUI1)
PHATRDRAFT_15371 uncharacterized membrane protein (bPH_2)
PHATRDRAFT_16807 zgc:162229 protein (Nudix_Hydrolase superfamily)
PHATRDRAFT_21441 mfs family transporter: sugar (MFS)
PHATRDRAFT_29423 conserved hypotheical protein (UPF0061 superfamily)
PHATRDRAFT_34422 family dioxygenase (TauD)
PHATRDRAFT_36969 cg9240-pa isoform 3 (S26_SPase_I)
PHATRDRAFT_44014 at1g22850 f29g20_19 (COG0398)
PHATRDRAFT_44076 ankyrin repeat
PHATRDRAFT_44224 (TRAM_LAG1_CLN8)
PHATRDRAFT_44303 2-oxoglutarate and iron-dependent oxygenase domain containing 2 (2OG-FeII_Oxy_3 superfamily)
PHATRDRAFT_45931 homo sapienskiaa0041 protein
PHATRDRAFT_46090 gcn5-related n-acetyltransferase (NAT_SF)
PHATRDRAFT_46273 (ubiquitin)
PHATRDRAFT_50545 (DUF2854 superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006413 | translational initiation | 0.0407909 | 1 | Phatr_bicluster_0156 |
| GO:0006508 | proteolysis and peptidolysis | 0.053216 | 1 | Phatr_bicluster_0156 |
| GO:0019538 | protein metabolism | 0.0606456 | 1 | Phatr_bicluster_0156 |
| GO:0006464 | protein modification | 0.104035 | 1 | Phatr_bicluster_0156 |
| GO:0016567 | protein ubiquitination | 0.118111 | 1 | Phatr_bicluster_0156 |
| GO:0006810 | transport | 0.287164 | 1 | Phatr_bicluster_0156 |

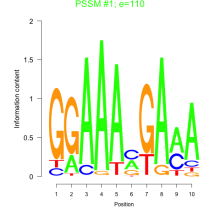

Comments