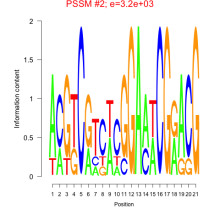
Phatr_bicluster_0162 Residual: 0.30
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0162 | 0.30 | Phaeodactylum tricornutum |
Displaying 1 - 36 of 36
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10187 (AdoMet_MTases)
PHATRDRAFT_10659 protein phosphatase 2c (PP2Cc)
PHATRDRAFT_15229 protein kinase (PKc_like superfamily)
PHATRDRAFT_15699 peroxisomal membrane (Mpv17_PMP22)
PHATRDRAFT_17115 trna-dihydrouridine synthase (DUS_like_FMN)
PHATRDRAFT_35989 jumonji domain containing 6 (JmjC superfamily)
PHATRDRAFT_38965 (VacB)
PHATRDRAFT_38974 set domain containing protein
PHATRDRAFT_41094 cg9924-isoform c (BTB)
PHATRDRAFT_47296 bernardinelli-seip congenital lipodystrophy 2 homologisoform cra_a (Seipin superfamily)
PHATRDRAFT_48341 uncharacterised conserved protein ucp028846 (DUF1237 superfamily)
PHATRDRAFT_48490 protein phosphatase 2c (PP2Cc)
PHATRDRAFT_49142 (Patatin_and_cPLA2 superfamily)
PHATRDRAFT_49280 zinc finger (c3hc4-type ring finger) family protein (RING)
PHATRDRAFT_49337 core 1 udp-galactose:n-acetylgalactosamine-alpha-r beta- galactosy
PHATRDRAFT_49419 mgc80481 protein
PHATRDRAFT_49500 non-ribosomal peptide synthetase (AFD_class_I superfamily)
PHATRDRAFT_4956 phosphoglycerate bisphosphoglycerate mutase family protein (His_Phos_1)
PHATRDRAFT_49898 methyltransferase type 11 (Glyco_trans_1_2)
PHATRDRAFT_50541 histone methyltransferase (SET)
PHATRDRAFT_5235 camp-dependent protein kinase regulatory subunit (CAP_ED)
PHATRDRAFT_5286 af435449_1calcium calmodulin-dependent protein kinase4 (S_TKc)
PHATRDRAFT_8461 adp-ribosylation factor gtpase activating protein 3 (ArfGap superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006468 | protein amino acid phosphorylation | 0.00232378 | 1 | Phatr_bicluster_0162 |
| GO:0043087 | regulation of GTPase activity | 0.0188766 | 1 | Phatr_bicluster_0162 |
| GO:0005975 | carbohydrate metabolism | 0.104035 | 1 | Phatr_bicluster_0162 |
| GO:0016567 | protein ubiquitination | 0.118111 | 1 | Phatr_bicluster_0162 |
| GO:0008152 | metabolism | 0.410263 | 1 | Phatr_bicluster_0162 |


Comments