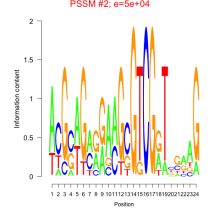
Phatr_bicluster_0166 Residual: 0.21
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0166 | 0.21 | Phaeodactylum tricornutum |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_32851 fructose-6-phosphate 2-kinase fructose--biphosphatase (AAA_17 superfamily)
PHATRDRAFT_34059 glycine oxidase (NAD_binding_8 superfamily)
PHATRDRAFT_35895 c1galt1-specific chaperone 1
PHATRDRAFT_44272 mgc114783 protein
PHATRDRAFT_45125 nadph-dependent fmn fad containing (FNR_like superfamily)
PHATRDRAFT_45267 deacetylase sulfotransferase
PHATRDRAFT_45348 4-hydroxybenzoate octaprenyltransferase (PT_UbiA superfamily)
PHATRDRAFT_45629 cupin 4 (Cupin_8 superfamily)
PHATRDRAFT_45910 ferredoxin (bacterial type ferredoxin family) (Fer4_15)
PHATRDRAFT_46742 d-lactate dehydrogenase (PRK11183)
PHATRDRAFT_47186 transcriptional repressor (NADB_Rossmann superfamily)
PHATRDRAFT_8970 mc family transporter
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006118 | electron transport | 0.00159697 | 1 | Phatr_bicluster_0166 |
| GO:0007218 | neuropeptide signaling pathway | 0.00246975 | 1 | Phatr_bicluster_0166 |
| GO:0006003 | fructose 2,6-bisphosphate metabolism | 0.004934 | 1 | Phatr_bicluster_0166 |
| GO:0000160 | two-component signal transduction system (phosphorelay) | 0.0624567 | 1 | Phatr_bicluster_0166 |
| GO:0006810 | transport | 0.38354 | 1 | Phatr_bicluster_0166 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.426896 | 1 | Phatr_bicluster_0166 |
| GO:0008152 | metabolism | 0.529843 | 1 | Phatr_bicluster_0166 |


Comments