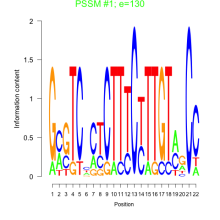
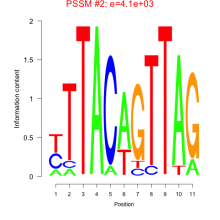
Phatr_bicluster_0173 Residual: 0.27
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0173 | 0.27 | Phaeodactylum tricornutum |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12262 peroxisomal membrane protein (Mpv17_PMP22)
PHATRDRAFT_13358 petc (photosynthetic electron transfer c) (PRK13474)
PHATRDRAFT_13706 long chain (FMN_red superfamily)
PHATRDRAFT_16421 (DUF791 superfamily)
PHATRDRAFT_32071 (DUF1517 superfamily)
PHATRDRAFT_34425 bass family transporter: sodium ion bile acid (SBF superfamily)
PHATRDRAFT_37719 acetamidase formamidase (FmdA_AmdA superfamily)
PHATRDRAFT_41845 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (lytB_ispH)
PHATRDRAFT_42494 large low complexity coiled coil protien with large repeat region
PHATRDRAFT_42788 (AdoMet_MTases superfamily)
PHATRDRAFT_43232 low-co2 inducible protein lcic
PHATRDRAFT_43593 lipase-like protein (Lipase_3)
PHATRDRAFT_44535 (Ion_trans)
PHATRDRAFT_47417 dna topoisomerase family protein (TopA)
PHATRDRAFT_48694 homoserine dehydrogenase (PRK06349)
PHATRDRAFT_54621 loc407614 protein (GT1_ALG11_like)
PHATRDRAFT_5928 zeaxanthin epoxidase (NAD_binding_8 superfamily)
PHATRDRAFT_9046 rieske (2fe-2s) region protein (Rieske_2)
PHATRDRAFT_9223 leu phe-trna protein (Leu_Phe_trans superfamily)
PHATRDRAFT_9799 light-harvesting complex i polypeptide (Chloroa_b-bind)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019288 | mevalonate-independent isopentenyl diphosphate biosynthesis | 0.00444554 | 1 | Phatr_bicluster_0173 |
| GO:0006118 | electron transport | 0.0162729 | 1 | Phatr_bicluster_0173 |
| GO:0006304 | DNA modification | 0.0220418 | 1 | Phatr_bicluster_0173 |
| GO:0006268 | DNA unwinding | 0.0220418 | 1 | Phatr_bicluster_0173 |
| GO:0008652 | amino acid biosynthesis | 0.0307293 | 1 | Phatr_bicluster_0173 |
| GO:0006265 | DNA topological change | 0.0393439 | 1 | Phatr_bicluster_0173 |
| GO:0006814 | sodium ion transport | 0.0478862 | 1 | Phatr_bicluster_0173 |
| GO:0030163 | protein catabolism | 0.0478862 | 1 | Phatr_bicluster_0173 |
| GO:0006811 | ion transport | 0.0478862 | 1 | Phatr_bicluster_0173 |
| GO:0006629 | lipid metabolism | 0.0772227 | 1 | Phatr_bicluster_0173 |

Comments