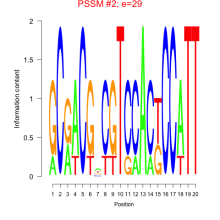
Phatr_bicluster_0224 Residual: 0.21
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0224 | 0.21 | Phaeodactylum tricornutum |
Displaying 1 - 18 of 18
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10800 (DUF819 superfamily)
PHATRDRAFT_11387 ca2+ h+ antiporter (cax)
PHATRDRAFT_15125 gdp-4-keto-6-deoxy-d-mannose epimerase-reductase (PLN02725)
PHATRDRAFT_18323 inositol-phosphate phosphatase (IMPase)
PHATRDRAFT_21006 protein kinase domain containing protein (Pkinase)
PHATRDRAFT_32849 branched-chain amino acid aminotransferase (BCAT_beta_family)
PHATRDRAFT_33037 (Methyltransf_28)
PHATRDRAFT_38394 3-ketoacyl-(acyl-carrier-protein) reductase (SDR_c)
PHATRDRAFT_44902 aspartate--ammonia ligase (PRK05425)
PHATRDRAFT_45495 two-component response regulator (PAS_9)
PHATRDRAFT_45572 ero1-like beta (ERO1 superfamily)
PHATRDRAFT_50964 o-acetylhomoserine o-acetylserine sulfhydrylase (CGS_like)
PHATRDRAFT_5780 3-ketoacyl-(acyl-carrier-protein) reductase (SDR_c)
PHATRDRAFT_8301 (DUF3593 superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009081 | branched chain family amino acid metabolism | 0.00591714 | 1 | Phatr_bicluster_0224 |
| GO:0006529 | asparagine biosynthesis | 0.00788271 | 1 | Phatr_bicluster_0224 |
| GO:0008152 | metabolism | 0.0161272 | 1 | Phatr_bicluster_0224 |
| GO:0006520 | amino acid metabolism | 0.0464863 | 1 | Phatr_bicluster_0224 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.0983212 | 1 | Phatr_bicluster_0224 |
| GO:0006468 | protein amino acid phosphorylation | 0.2936 | 1 | Phatr_bicluster_0224 |


Comments