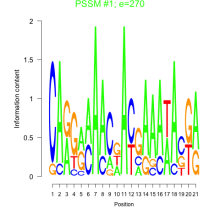
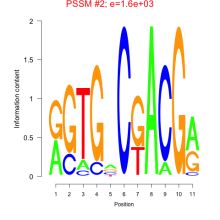
Phatr_bicluster_0256 Residual: 0.28
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0256 | 0.28 | Phaeodactylum tricornutum |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_16074 non-specific dna-binding protein (HU)
PHATRDRAFT_17198 isoleucyl-trna synthetase (PLN02843)
PHATRDRAFT_17332 band 7 protein (SPFH_paraslipin)
PHATRDRAFT_19690 fkbp-type peptidyl-prolyl cis-trans isomerase (FKBP_C)
PHATRDRAFT_21116 argininosuccinate synthetase (Argininosuccinate_Synthase)
PHATRDRAFT_25420 rna polymeraseii and iia subunit (PLN03111)
PHATRDRAFT_26346 cysteinyl-trna synthetase (PTZ00399)
PHATRDRAFT_31768 elongation factor ts (tsf)
PHATRDRAFT_32846 (Methyltrans_RNA superfamily)
PHATRDRAFT_37579 mitochondrial ribosomal (Nudix_Hydrolase superfamily)
PHATRDRAFT_42754 (RRM_SF)
PHATRDRAFT_45039 glycosyl transferase (Gly_transf_sug superfamily)
PHATRDRAFT_46163 phytanoyl-dioxygenase family protein (PhyH superfamily)
PHATRDRAFT_46320 eif3a (eukaryotic translation initiation factor 3a)
PHATRDRAFT_46461 homoserine (PTZ00299)
PHATRDRAFT_46522 (NTF2)
PHATRDRAFT_47116 polymeraseiii (dna directed) polypeptide c
PHATRDRAFT_47412 (DEXDc)
PHATRDRAFT_47492 mgc68780 protein (TPR)
PHATRDRAFT_47999 ac007980_9cytosolic trna-ala synthetase (PLN02900)
PHATRDRAFT_51005 ligase i (PRK01109)
PHATRDRAFT_51868 amine oxidase (NAD_binding_8 superfamily)
PHATRDRAFT_8720 50s ribosomal protein l17 (rplQ)
PHATRDRAFT_9197 h aca ribonucleoprotein complex subunit 3 (Nop10p)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006350 | transcription | 0.00332513 | 1 | Phatr_bicluster_0256 |
| GO:0006566 | threonine metabolism | 0.00419857 | 1 | Phatr_bicluster_0256 |
| GO:0006428 | isoleucyl-tRNA aminoacylation | 0.00838054 | 1 | Phatr_bicluster_0256 |
| GO:0006423 | cysteinyl-tRNA aminoacylation | 0.012546 | 1 | Phatr_bicluster_0256 |
| GO:0006606 | protein-nucleus import | 0.012546 | 1 | Phatr_bicluster_0256 |
| GO:0006419 | alanyl-tRNA aminoacylation | 0.012546 | 1 | Phatr_bicluster_0256 |
| GO:0006526 | arginine biosynthesis | 0.0290436 | 1 | Phatr_bicluster_0256 |
| GO:0016310 | phosphorylation | 0.057292 | 1 | Phatr_bicluster_0256 |
| GO:0006414 | translational elongation | 0.057292 | 1 | Phatr_bicluster_0256 |
| GO:0006260 | DNA replication | 0.103926 | 1 | Phatr_bicluster_0256 |

Comments