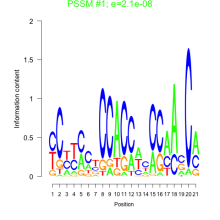
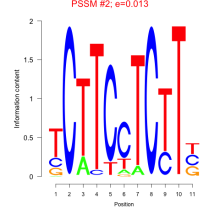
Thaps_bicluster_0033 Residual: 0.48
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0033 | 0.48 | Thalassiosira pseudonana |
Displaying 1 - 47 of 47
" class="views-fluidgrid-wrapper clear-block">
1077 hypothetical protein
10848 hypothetical protein
11516 hypothetical protein
11592 hypothetical protein
11801 Methyltrans_RNA superfamily
12164 hypothetical protein
16656 MPP_Nbla03831
1751 hypothetical protein
1797 hypothetical protein
18190 P4Hc
19479 PhyH superfamily
19890 CbpA
20688 (Tp_CBF/NF5) regulator [Rayko]
20834 DLP_2
22643 hypothetical protein
22950 DUF1501 superfamily
24888 hypothetical protein
261219 Mad3_BUB1_I superfamily
261384 niemann-pick C type protein-like protein
262335 hypothetical protein
262514 (HAL3) Flavoprotein superfamily
262580 AdoMet_dc
262854 CBS_pair superfamily
263700 HA
264902 (cht1) chitinase
270228 (Lhcx4) fucoxanthin chlorophyll a/c protein, LI818 clade
27435 PAH
2808 PKc_like superfamily
31142 H15
31174 hypothetical protein
31803 MYSc
33055 PLN02256
34233 AdoMet_dc
35059 MYSc
35186 PRK13357
35499 Smc
3607 hypothetical protein
3660 COG5038
3788 Methyltransf_22 superfamily
38514 DUF3593
4590 hypothetical protein
4915 hypothetical protein
5317 hypothetical protein
5551 hypothetical protein
5722 hypothetical protein
6404 hypothetical protein
9691 LRR_RI superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009081 | branched chain family amino acid metabolism | 0.00578093 | 1 | Thaps_bicluster_0033 |
| GO:0051276 | chromosome organization and biogenesis | 0.0143942 | 1 | Thaps_bicluster_0033 |
| GO:0016567 | protein ubiquitination | 0.0211078 | 1 | Thaps_bicluster_0033 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0546807 | 1 | Thaps_bicluster_0033 |
| GO:0019538 | protein metabolism | 0.125374 | 1 | Thaps_bicluster_0033 |
| GO:0006457 | protein folding | 0.317461 | 1 | Thaps_bicluster_0033 |
| GO:0008152 | metabolism | 0.332159 | 1 | Thaps_bicluster_0033 |
| GO:0006468 | protein amino acid phosphorylation | 0.468128 | 1 | Thaps_bicluster_0033 |
| GO:0006118 | electron transport | 0.547503 | 1 | Thaps_bicluster_0033 |
| GO:0006508 | proteolysis and peptidolysis | 0.577098 | 1 | Thaps_bicluster_0033 |

Comments