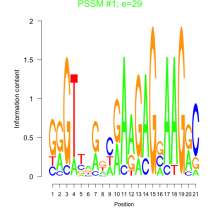
Thaps_bicluster_0103 Residual: 0.42
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0103 | 0.42 | Thalassiosira pseudonana |
Displaying 1 - 38 of 38
" class="views-fluidgrid-wrapper clear-block">
10346 hypothetical protein
10363 SET
11142 hypothetical protein
11176 MPP_CSTP1
11193 DUF159
11319 hypothetical protein
17242 SPFH_like_u4
1932 HA
19977 TRX_family
21362 hypothetical protein
21720 hypothetical protein
22016 ABC_MTABC3_MDL1_MDL2
22155 hypothetical protein
22297 VPS9 superfamily
22430 SOUL
22644 hypothetical protein
23409 hypothetical protein
23771 hypothetical protein
24456 hypothetical protein
24659 DnaQ_like_exo superfamily
24738 hypothetical protein
24971 Tcp11 superfamily
25102 hypothetical protein
268460 PolY superfamily
31845 PLN02428
32176 GABARAP
33480 DUF924
36015 hypothetical protein
37995 DUF2838
3953 hypothetical protein
4395 hypothetical protein
5388 hypothetical protein
6396 Dabb
6476 hypothetical protein
6723 hypothetical protein
7268 ATPase-IIIA_H
7667 hypothetical protein
7889 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009107 | lipoate biosynthesis | 0.00742418 | 1 | Thaps_bicluster_0103 |
| GO:0015992 | proton transport | 0.0275887 | 1 | Thaps_bicluster_0103 |
| GO:0006813 | potassium ion transport | 0.0402303 | 1 | Thaps_bicluster_0103 |
| GO:0006812 | cation transport | 0.0772831 | 1 | Thaps_bicluster_0103 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.0824713 | 1 | Thaps_bicluster_0103 |
| GO:0006281 | DNA repair | 0.0824713 | 1 | Thaps_bicluster_0103 |
| GO:0006810 | transport | 0.294845 | 1 | Thaps_bicluster_0103 |
| GO:0006118 | electron transport | 0.399203 | 1 | Thaps_bicluster_0103 |
| GO:0008152 | metabolism | 0.546944 | 1 | Thaps_bicluster_0103 |


Comments