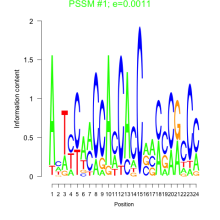
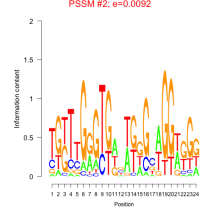
Thaps_bicluster_0107 Residual: 0.36
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0107 | 0.36 | Thalassiosira pseudonana |
Displaying 1 - 26 of 26
" class="views-fluidgrid-wrapper clear-block">
11338 hypothetical protein
1481 hypothetical protein
17910 UBCc
20951 ARM
21415 hypothetical protein
21581 hypothetical protein
22118 Pkinase
23825 hypothetical protein
23899 hypothetical protein
23946 hypothetical protein
24068 SNARE component
24142 hypothetical protein
24719 hypothetical protein
25378 hypothetical protein
25440 Smc
263031 (Rad23) rad23
264095 (TF2D2) PLN00062
34620 PX_SNX1_2_like
35907 Prp18
36524 Elf1
37430 Sm_D2
6600 Sm_D1
6704 hypothetical protein
7791 hypothetical protein
7903 hypothetical protein
9303 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006397 | mRNA processing | 0.0014945 | 1 | Thaps_bicluster_0107 |
| GO:0008380 | RNA splicing | 0.00536856 | 1 | Thaps_bicluster_0107 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 0.00536856 | 1 | Thaps_bicluster_0107 |
| GO:0006464 | protein modification | 0.0104659 | 1 | Thaps_bicluster_0107 |
| GO:0006289 | nucleotide-excision repair | 0.0133715 | 1 | Thaps_bicluster_0107 |
| GO:0006350 | transcription | 0.0777882 | 1 | Thaps_bicluster_0107 |
| GO:0006512 | ubiquitin cycle | 0.0926592 | 1 | Thaps_bicluster_0107 |
| GO:0007242 | intracellular signaling cascade | 0.107309 | 1 | Thaps_bicluster_0107 |
| GO:0006886 | intracellular protein transport | 0.135954 | 1 | Thaps_bicluster_0107 |
| GO:0006468 | protein amino acid phosphorylation | 0.443559 | 1 | Thaps_bicluster_0107 |

Comments