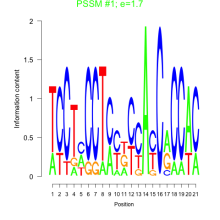
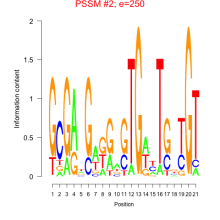
Thaps_bicluster_0141 Residual: 0.18
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0141 | 0.18 | Thalassiosira pseudonana |
Displaying 1 - 12 of 12
" class="views-fluidgrid-wrapper clear-block">
12971 WD40 superfamily
22441 hypothetical protein
22499 TrmA
23210 hypothetical protein
23336 SRP40_C superfamily
25120 U3_assoc_6 superfamily
261145 Lactamase_B superfamily
262018 hypothetical protein
32158 NOC3
34021 Methyltrn_RNA_3
34176 AdoMet_MTases superfamily
38662 BING4CT
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006310 | DNA recombination | 0.0117404 | 1 | Thaps_bicluster_0141 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.0349447 | 1 | Thaps_bicluster_0141 |
| GO:0006396 | RNA processing | 0.0355504 | 1 | Thaps_bicluster_0141 |

Comments