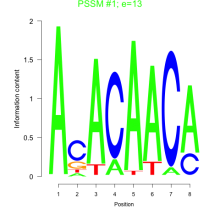
Thaps_bicluster_0169 Residual: 0.34
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0169 | 0.34 | Thalassiosira pseudonana |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
10102 hypothetical protein
10280 hypothetical protein
11506 hypothetical protein
13134 AarF
21489 Y_phosphatase3
22388 hypothetical protein
22880 Fasciclin
23931 PLN02244
24285 hypothetical protein
24555 hypothetical protein
270329 hypothetical protein
2790 PLN02291
30895 Aldo_ket_red
3298 MtN3_slv superfamily
34399 FolD
3854 hypothetical protein
39520 hppA
4065 hypothetical protein
4452 CDC14
4767 hypothetical protein
5389 hypothetical protein
6896 Chalcone superfamily
7035 hypothetical protein
7474 PITH
7957 DUF3464
9394 hypothetical protein
974 hot_dog superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009396 | folic acid and derivative biosynthesis | 0.0129601 | 1 | Thaps_bicluster_0169 |
| GO:0009073 | aromatic amino acid family biosynthesis | 0.0129601 | 1 | Thaps_bicluster_0169 |
| GO:0015992 | proton transport | 0.0275887 | 1 | Thaps_bicluster_0169 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0420242 | 1 | Thaps_bicluster_0169 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0473882 | 1 | Thaps_bicluster_0169 |
| GO:0007155 | cell adhesion | 0.0544987 | 1 | Thaps_bicluster_0169 |
| GO:0006468 | protein amino acid phosphorylation | 0.333459 | 1 | Thaps_bicluster_0169 |
| GO:0006508 | proteolysis and peptidolysis | 0.424754 | 1 | Thaps_bicluster_0169 |
| GO:0008152 | metabolism | 0.546944 | 1 | Thaps_bicluster_0169 |


Comments