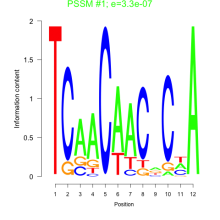
Thaps_bicluster_0183 Residual: 0.30
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0183 | 0.30 | Thalassiosira pseudonana |
Displaying 1 - 20 of 20
" class="views-fluidgrid-wrapper clear-block">
10241 hypothetical protein
1506 hypothetical protein
18469 FkpA
1969 hypothetical protein
21250 hypothetical protein
22117 hypothetical protein
22140 UFD1 superfamily
22418 hypothetical protein
269968 hypothetical protein
270261 hypothetical protein
3257 hypothetical protein
33389 PKc_like superfamily
36527 hypothetical protein
36557 PAP_fibrillin superfamily
4722 hypothetical protein
4876 hypothetical protein
4888 hypothetical protein
5545 hypothetical protein
6807 hypothetical protein
8793 DUF328
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.0415252 | 1 | Thaps_bicluster_0183 |
| GO:0006457 | protein folding | 0.103279 | 1 | Thaps_bicluster_0183 |
| GO:0006468 | protein amino acid phosphorylation | 0.164893 | 1 | Thaps_bicluster_0183 |
| GO:0006508 | proteolysis and peptidolysis | 0.217787 | 1 | Thaps_bicluster_0183 |


Comments