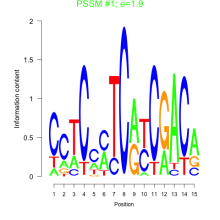
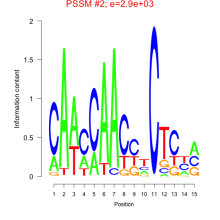
Thaps_bicluster_0206 Residual: 0.21
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0206 | 0.21 | Thalassiosira pseudonana |
Displaying 1 - 16 of 16
" class="views-fluidgrid-wrapper clear-block">
10133 hypothetical protein
10286 hypothetical protein
10295 hypothetical protein
1054 hypothetical protein
10641 hypothetical protein
1305 hypothetical protein
2053 hypothetical protein
264191 WD40 superfamily
36420 PLN03128
3882 hypothetical protein
4653 hypothetical protein
6004 hypothetical protein
7024 PLDc_PMFPLD_like_1
8926 MDR8
9367 hypothetical protein
9764 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006265 | DNA topological change | 0.0102986 | 1 | Thaps_bicluster_0206 |
| GO:0006259 | DNA metabolism | 0.0143942 | 1 | Thaps_bicluster_0206 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0235597 | 1 | Thaps_bicluster_0206 |
| GO:0008152 | metabolism | 0.0595321 | 1 | Thaps_bicluster_0206 |

Comments