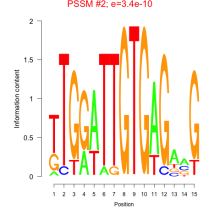
Thaps_bicluster_0207 Residual: 0.30
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0207 | 0.30 | Thalassiosira pseudonana |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
10181 hypothetical protein
11420 hypothetical protein
11559 hypothetical protein
12162 hypothetical protein
12168 hypothetical protein
12193 hypothetical protein
12196 hypothetical protein
12197 hypothetical protein
23656 (Tp_AP2-EREBP7) regulator [Rayko]
26082 SRP1
31391 KISc
3183 PTZ00018
33270 PLN00157
34210 PLN00158
35279 (H4_2) H4
36811 PTZ00018
37309 PLN00158
37357 H4
37431 PLN00157
37432 PTZ00018
4261 hypothetical protein
5018 STKc_MAPK
6947 PLN00158
6948 PLN00157
9067 (H4_1) H4
9991 hypothetical protein
9992 H15
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006334 | nucleosome assembly | 8.47e-25 | 2.35e-21 | Thaps_bicluster_0207 |
| GO:0007001 | chromosome organization and biogenesis (sensu Eukaryota) | 6.15e-23 | 1.71e-19 | Thaps_bicluster_0207 |
| GO:0007018 | microtubule-based movement | 0.235625 | 1 | Thaps_bicluster_0207 |
| GO:0006468 | protein amino acid phosphorylation | 0.742103 | 1 | Thaps_bicluster_0207 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.861003 | 1 | Thaps_bicluster_0207 |


Comments