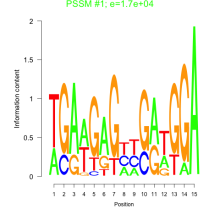
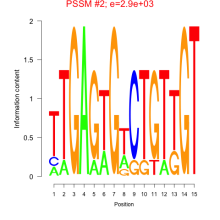
Thaps_bicluster_0210 Residual: 0.31
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0210 | 0.31 | Thalassiosira pseudonana |
Displaying 1 - 19 of 19
" class="views-fluidgrid-wrapper clear-block">
11111 DUF383 superfamily
20960 hypothetical protein
21422 hypothetical protein
21971 hypothetical protein
22045 (Tp_CCHH10) regulator [Rayko]
2237 hypothetical protein
23300 hypothetical protein
23490 Alpha_kinase
262731 hypothetical protein
263046 HA
263463 (Tp_Hox6b) regulator [Rayko]
3315 hypothetical protein
34357 Fcf2
35584 AANH_like superfamily
38361 HMGB-UBF_HMG-box
5387 hypothetical protein
7838 hypothetical protein
8559 hypothetical protein
9045 TLC superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006952 | defense response | 0.00247959 | 1 | Thaps_bicluster_0210 |
| GO:0006350 | transcription | 0.0366595 | 1 | Thaps_bicluster_0210 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0508003 | 1 | Thaps_bicluster_0210 |
| GO:0006810 | transport | 0.207702 | 1 | Thaps_bicluster_0210 |
| GO:0006468 | protein amino acid phosphorylation | 0.236888 | 1 | Thaps_bicluster_0210 |

Comments