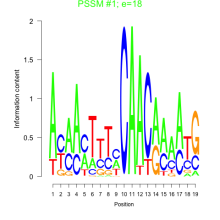
Thaps_bicluster_0212 Residual: 0.16
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0212 | 0.16 | Thalassiosira pseudonana |
Displaying 1 - 13 of 13
" class="views-fluidgrid-wrapper clear-block">
10180 hypothetical protein
1052 hypothetical protein
24064 hypothetical protein
25912 hypothetical protein
261036 PLN03150
33883 CYCLIN
3482 hypothetical protein
4771 hypothetical protein
6330 hypothetical protein
6699 hypothetical protein
6773 hypothetical protein
6774 hypothetical protein
7087 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0000074 | regulation of cell cycle | 0.0124044 | 1 | Thaps_bicluster_0212 |


Comments