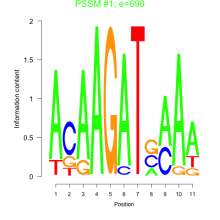
Thaps_bicluster_0215 Residual: 0.25
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0215 | 0.25 | Thalassiosira pseudonana |
Displaying 1 - 17 of 17
" class="views-fluidgrid-wrapper clear-block">
11183 hypothetical protein
23516 hypothetical protein
25077 hypothetical protein
262575 DHOD_1A_like
264628 UxaC
268773 Gst
2698 hypothetical protein
3322 hypothetical protein
33950 TMEM18
34684 Trypsin_2
4292 hypothetical protein
5098 hypothetical protein
5872 hypothetical protein
5971 hypothetical protein
6757 hypothetical protein
8824 hypothetical protein
9795 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006207 | 'de novo' pyrimidine base biosynthesis | 0.00206654 | 1 | Thaps_bicluster_0215 |
| GO:0006508 | proteolysis and peptidolysis | 0.115549 | 1 | Thaps_bicluster_0215 |


Comments