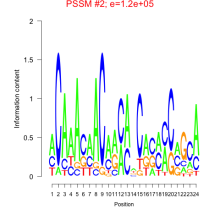
Thaps_bicluster_0216 Residual: 0.36
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0216 | 0.36 | Thalassiosira pseudonana |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
11710 hypothetical protein
1194 hypothetical protein
19158 GRX_PICOT_like
21451 hypothetical protein
2243 hypothetical protein
22481 hypothetical protein
22783 CRCB
23394 GAT_1 superfamily
23786 DUF547 superfamily
23841 hypothetical protein
24199 hypothetical protein
24961 hypothetical protein
25106 hypothetical protein
25322 FlaRed superfamily
25449 hypothetical protein
25450 hypothetical protein
25608 hypothetical protein
25626 hypothetical protein
25901 MviM
262597 (FRU2) hypothetical protein
264399 YafJ
264918 2A0303
268064 (Tp_HSF_2.7d) HSF_DNA-bind superfamily
268481 (Tp_HSF_2.2a) HSF_DNA-bind
268516 hypothetical protein
268653 hypothetical protein
2877 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0045454 | cell redox homeostasis | 0.000319158 | 0.870345 | Thaps_bicluster_0216 |
| GO:0006118 | electron transport | 0.0457659 | 1 | Thaps_bicluster_0216 |
| GO:0006508 | proteolysis and peptidolysis | 0.0557851 | 1 | Thaps_bicluster_0216 |
| GO:0006865 | amino acid transport | 0.0863737 | 1 | Thaps_bicluster_0216 |
| GO:0016567 | protein ubiquitination | 0.219149 | 1 | Thaps_bicluster_0216 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.245811 | 1 | Thaps_bicluster_0216 |
| GO:0008152 | metabolism | 0.363828 | 1 | Thaps_bicluster_0216 |
| GO:0006810 | transport | 0.441554 | 1 | Thaps_bicluster_0216 |


Comments