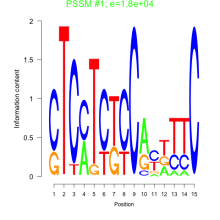
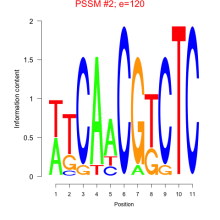
Thaps_bicluster_0220 Residual: 0.24
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0220 | 0.24 | Thalassiosira pseudonana |
Displaying 1 - 15 of 15
" class="views-fluidgrid-wrapper clear-block">
20847 hypothetical protein
23875 hypothetical protein
24160 hypothetical protein
24408 hypothetical protein
25464 hypothetical protein
260926 PLN00113
3123 hypothetical protein
3361 SbcC
3646 hypothetical protein
39677 (Tp_Myb3R4) regulator [Rayko]
4300 SCP superfamily
6203 Bestrophin superfamily
6580 hypothetical protein
7023 hypothetical protein
7236 hypothetical protein

Comments