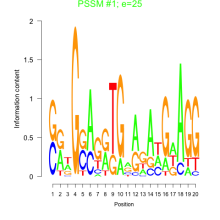
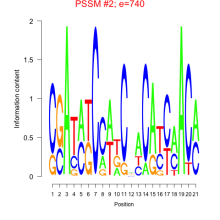
Thaps_bicluster_0231 Residual: 0.32
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0231 | 0.32 | Thalassiosira pseudonana |
Displaying 1 - 21 of 21
" class="views-fluidgrid-wrapper clear-block">
10302 hypothetical protein
11738 hypothetical protein
11831 hypothetical protein
22963 WD40 superfamily
23341 hypothetical protein
2462 hypothetical protein
25886 DUF1824 superfamily
263200 PLN02605
264925 Flavodoxin_1
268844 hypothetical protein
2950 (UroS) HemD
32874 SodA
37838 M32_Taq
40655 PRK06349
4160 PMT_2 superfamily
4225 hypothetical protein
6942 DUF89
7160 hypothetical protein
8872 hypothetical protein
9166 hypothetical protein
9776 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006486 | protein amino acid glycosylation | 0.00557274 | 1 | Thaps_bicluster_0231 |
| GO:0006801 | superoxide metabolism | 0.00742418 | 1 | Thaps_bicluster_0231 |
| GO:0006783 | heme biosynthesis | 0.00742418 | 1 | Thaps_bicluster_0231 |
| GO:0008652 | amino acid biosynthesis | 0.0202987 | 1 | Thaps_bicluster_0231 |
| GO:0006779 | porphyrin biosynthesis | 0.0202987 | 1 | Thaps_bicluster_0231 |
| GO:0009058 | biosynthesis | 0.0978804 | 1 | Thaps_bicluster_0231 |
| GO:0006118 | electron transport | 0.399203 | 1 | Thaps_bicluster_0231 |
| GO:0006508 | proteolysis and peptidolysis | 0.424754 | 1 | Thaps_bicluster_0231 |
| GO:0008152 | metabolism | 0.546944 | 1 | Thaps_bicluster_0231 |

Comments