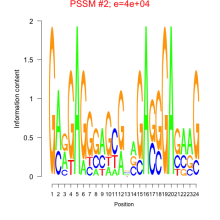
Thaps_bicluster_0248 Residual: 0.34
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0248 | 0.34 | Thalassiosira pseudonana |
Displaying 1 - 33 of 33
" class="views-fluidgrid-wrapper clear-block">
10006 UCH
1322 hypothetical protein
17841 RNase_PH superfamily
19136 hypothetical protein
1943 (Tp_HSF_2.6e) HSF_DNA-bind superfamily
2050 hypothetical protein
20579 hypothetical protein
21537 hypothetical protein
22258 PUB
24091 choice_anch_B
24130 hypothetical protein
24721 hypothetical protein
25030 hypothetical protein
25048 hypothetical protein
25658 hypothetical protein
261392 NPR2 superfamily
262516 hypothetical protein
262797 Sec7
263000 RRM2_U2AF65
263108 Smc
263124 RRM_snRNP70
263184 lysozyme_like superfamily
263198 Cob_adeno_trans
264371 MFS
268044 hypothetical protein
269773 hypothetical protein
2912 hypothetical protein
34838 UBCc superfamily
42283 RabGAP-TBC
5854 hypothetical protein
5858 hypothetical protein
6628 hypothetical protein
7982 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015904 | tetracycline transport | 0.0143942 | 1 | Thaps_bicluster_0248 |
| GO:0006032 | chitin catabolism | 0.0229381 | 1 | Thaps_bicluster_0248 |
| GO:0006364 | rRNA processing | 0.0229381 | 1 | Thaps_bicluster_0248 |
| GO:0016998 | cell wall catabolism | 0.0257707 | 1 | Thaps_bicluster_0248 |
| GO:0009613 | response to pest, pathogen or parasite | 0.0257707 | 1 | Thaps_bicluster_0248 |
| GO:0006350 | transcription | 0.0835234 | 1 | Thaps_bicluster_0248 |
| GO:0006512 | ubiquitin cycle | 0.0994304 | 1 | Thaps_bicluster_0248 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.138083 | 1 | Thaps_bicluster_0248 |
| GO:0006396 | RNA processing | 0.155589 | 1 | Thaps_bicluster_0248 |
| GO:0006464 | protein modification | 0.158063 | 1 | Thaps_bicluster_0248 |


Comments