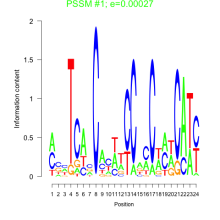
Thaps_bicluster_0253 Residual: 0.34
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0253 | 0.34 | Thalassiosira pseudonana |
Displaying 1 - 31 of 31
" class="views-fluidgrid-wrapper clear-block">
10836 hypothetical protein
11048 hypothetical protein
11394 hypothetical protein
11932 hypothetical protein
11933 hypothetical protein
1764 hypothetical protein
1976 hypothetical protein
21636 hypothetical protein
22701 hypothetical protein
23603 hypothetical protein
24561 hypothetical protein
24894 hypothetical protein
2574 hypothetical protein
262248 proline-rich protein
262249 hypothetical protein
262378 (FRU1) hypothetical protein
263215 SCP superfamily
2635 RCC1
263936 Peptidases_S8_Kp43_protease
264656 Tryp_SPc
4229 hypothetical protein
4668 hypothetical protein
4781 hypothetical protein
4796 hypothetical protein
5120 hypothetical protein
5167 hypothetical protein
5357 hypothetical protein
6944 hypothetical protein
8705 hypothetical protein
9440 hypothetical protein
9557 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006508 | proteolysis and peptidolysis | 0.0101826 | 1 | Thaps_bicluster_0253 |
| GO:0006457 | protein folding | 0.0784967 | 1 | Thaps_bicluster_0253 |


Comments