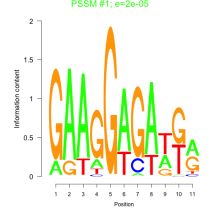
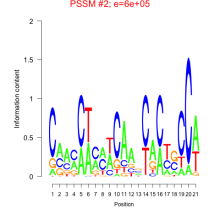
Thaps_bicluster_0266 Residual: 0.39
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0266 | 0.39 | Thalassiosira pseudonana |
Displaying 1 - 37 of 37
" class="views-fluidgrid-wrapper clear-block">
10325 hypothetical protein
10501 (DES2) Delta6-FADS-like
10955 hypothetical protein
10989 hypothetical protein
11055 hypothetical protein
11425 hypothetical protein
11510 hypothetical protein
11688 hypothetical protein
11712 hypothetical protein
11743 hypothetical protein
11965 hypothetical protein
2052 hypothetical protein
20752 hypothetical protein
21388 hypothetical protein
22571 hypothetical protein
24455 hypothetical protein
260838 Trichoplein
262112 ANK
2797 hypothetical protein
33437 YjgF_YER057c_UK114_like_2
3415 hypothetical protein
36664 hypothetical protein
3979 hypothetical protein
4071 hypothetical protein
4555 hypothetical protein
4969 hypothetical protein
5602 hypothetical protein
5838 hypothetical protein
5970 DUF1619 superfamily
6341 CEP19
6344 hypothetical protein
6633 hypothetical protein
6637 hypothetical protein
7378 leucine-rich repeat protein
7999 hypothetical protein
9079 IFT57
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006636 | fatty acid desaturation | 0.0123481 | 1 | Thaps_bicluster_0266 |
| GO:0007160 | cell-matrix adhesion | 0.0184762 | 1 | Thaps_bicluster_0266 |
| GO:0006725 | aromatic compound metabolism | 0.0225447 | 1 | Thaps_bicluster_0266 |
| GO:0006306 | DNA methylation | 0.0296321 | 1 | Thaps_bicluster_0266 |
| GO:0007155 | cell adhesion | 0.0306412 | 1 | Thaps_bicluster_0266 |

Comments