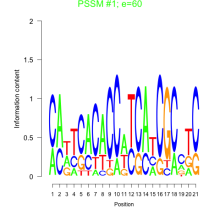
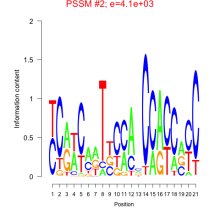
Thaps_bicluster_0270 Residual: 0.28
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0270 | 0.28 | Thalassiosira pseudonana |
Displaying 1 - 15 of 15
" class="views-fluidgrid-wrapper clear-block">
15424 hypothetical protein
17110 rbgA
20580 hypothetical protein
20626 hypothetical protein
20629 YCII superfamily
22221 hypothetical protein
23385 hypothetical protein
268238 PRK05192
268350 SpoU_methylase
2920 PRK05764
30691 AdoMet_MTases
35224 Aldo_ket_red
36186 iojap_ybeB
40223 PLN02347
6780 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0000154 | rRNA modification | 0.00454357 | 1 | Thaps_bicluster_0270 |
| GO:0006177 | GMP biosynthesis | 0.00454357 | 1 | Thaps_bicluster_0270 |
| GO:0009058 | biosynthesis | 0.00653848 | 1 | Thaps_bicluster_0270 |
| GO:0006164 | purine nucleotide biosynthesis | 0.00906837 | 1 | Thaps_bicluster_0270 |
| GO:0006310 | DNA recombination | 0.0424129 | 1 | Thaps_bicluster_0270 |
| GO:0006260 | DNA replication | 0.0768533 | 1 | Thaps_bicluster_0270 |
| GO:0006281 | DNA repair | 0.0998737 | 1 | Thaps_bicluster_0270 |
| GO:0006396 | RNA processing | 0.124391 | 1 | Thaps_bicluster_0270 |
| GO:0006118 | electron transport | 0.463588 | 1 | Thaps_bicluster_0270 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.514255 | 1 | Thaps_bicluster_0270 |

Comments