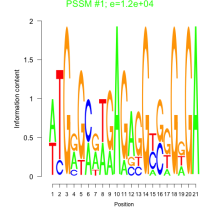
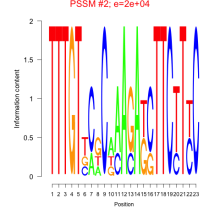
Thaps_bicluster_0282 Residual: 0.29
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0282 | 0.29 | Thalassiosira pseudonana |
Displaying 1 - 33 of 33
" class="views-fluidgrid-wrapper clear-block">
10458 hypothetical protein
11138 hypothetical protein
11150 hypothetical protein
11499 hypothetical protein
11817 hypothetical protein
1940 hypothetical protein
20891 Cyt_b561_FRRS1_like
21595 hypothetical protein
22062 hypothetical protein
22281 hypothetical protein
22630 hypothetical protein
22631 hypothetical protein
23757 hypothetical protein
24312 hypothetical protein
25038 hypothetical protein
25408 hypothetical protein
25838 hypothetical protein
25913 hypothetical protein
25921 (TPSIL2) silaffin protein
262856 Tryp_SPc
3192 NADB_Rossmann superfamily
33337 CAP_ED
3424 hypothetical protein
4495 hypothetical protein
4782 hypothetical protein
4922 Peptidases_S8_Kp43_protease
5439 hypothetical protein
7900 SCP superfamily
8086 hypothetical protein
8273 hypothetical protein
8927 hypothetical protein
9432 SCP superfamily
9434 Tryp_SPc
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0000074 | regulation of cell cycle | 0.00515106 | 1 | Thaps_bicluster_0282 |
| GO:0006508 | proteolysis and peptidolysis | 0.0134124 | 1 | Thaps_bicluster_0282 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.0824713 | 1 | Thaps_bicluster_0282 |
| GO:0008152 | metabolism | 0.171652 | 1 | Thaps_bicluster_0282 |
| GO:0006468 | protein amino acid phosphorylation | 0.333459 | 1 | Thaps_bicluster_0282 |

Comments