MMP1186 MMP1186 Eukaryotic thiol (cysteine) protease, active site:ATP/GTP-binding site motif A (P-loop):Sigma-54 factor interaction domain:Pu... (NCBI)
| Locus Tag | Symbol | Synonyms | Product | Protein Synonyms | Start | End | Strand | PubMed ID | |
|---|---|---|---|---|---|---|---|---|---|
| MMP1186 | MMP1186 | lon | lon, MMP1186 | Eukaryotic thiol (cysteine) protease, active site:ATP/GTP-binding site motif A (P-loop):Sigma-54... | Lon protein (EC 3.4.21.53) | 1168587 | 1170665 | + | 15466049 |
| Residual | Motif 1 | Expression Plot | |
|---|---|---|---|
| mmp-bicluster_0021 | 0.45 |
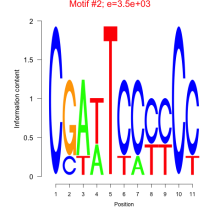 |
|
| mmp-bicluster_0115 | 0.49 |
 |
| Line | Position | Effect | Impact | Class | Percent Frequency | Sample | Title |
|---|---|---|---|---|---|---|---|
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 99 | HA2.152.08.M01 | BX950229-MMP1186-1169640-HA2.152.08.M01 | |
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 99 | HA2.152.01.M01 | BX950229-MMP1186-1169640-HA2.152.01.M01 | |
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 100 | HA2.152.01.M03 | BX950229-MMP1186-1169640-HA2.152.01.M03 | |
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 99 | HA2.152.01.M02 | BX950229-MMP1186-1169640-HA2.152.01.M02 | |
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 100 | HA2.152.05.M03 | BX950229-MMP1186-1169640-HA2.152.05.M03 | |
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 100 | HA2.152.09.M02 | BX950229-MMP1186-1169640-HA2.152.09.M02 | |
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 96 | HA2.152.08.M03 | BX950229-MMP1186-1169640-HA2.152.08.M03 | |
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 100 | HA2.152.05.M02 | BX950229-MMP1186-1169640-HA2.152.05.M02 | |
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 100 | HA2.152.09.M01 | BX950229-MMP1186-1169640-HA2.152.09.M01 | |
| HA2 | 1 169 640 | NON_SYNONYMOUS_CODING | MISSENSE | 100 | HA2.152.09.M03 | BX950229-MMP1186-1169640-HA2.152.09.M03 |
| Title | Insert | TAG Module |
|---|---|---|
| MMP1186 |
| Orthologues | Paralogues |
|---|---|
| COG | EC Description | TIGR Roles |
|---|---|---|
| (O) COG1067 | Predicted ATP-dependent protease | (3.4.21.53) Endopeptidase La. | Protein fate:Degradation of proteins, peptides, and glycopeptides |
| L1.Cm.T1 | L1.Cm.T2 | L1.cN.T1 | L1.cN.T2 | L1.T0 | L2.Cm.T1 | L2.Cm.T2 | L2.cN.T1 | L2.cN.T2 | L2.T0 |
|---|---|---|---|---|---|---|---|---|---|
| 3 | 1 | 6 | 0 | 32 | 3 | 5 | 4 | 2 | 22 |
| Product (LegacyBRC) | Product (RefSeq) |
|---|---|
| GI Number | Accession | Blast | Conserved Domains |
|---|---|---|---|
| 45358749 | NP_988306.1 | Run |
| Link to STRINGS | STRINGS Network |
|---|---|
| MMP1186 |

Add comment