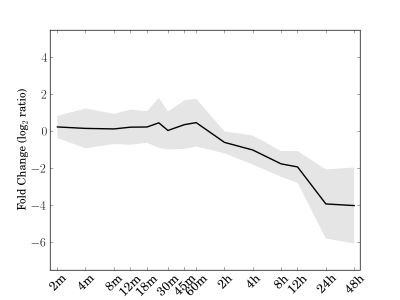
Module 133 Residual: 0.21
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0133 | v02 | 0.21 | -16.16 |
Displaying 1 - 16 of 16
| Cre01.g031250.t1.2 Protein of unknown function (DUF1279) | Cre03.g194450.t1.1 Predicted protein |
| Cre04.g223100.t1.2 alpha carbonic anhydrase 4 | Cre05.g248600.t1.2 P-type ATPase of Arabidopsis 2 |
| Cre09.g391208.t1.1 Plastid-specific ribosomal protein 4 | Cre11.g467572.t2.1 |
| Cre12.g484150.t1.1 SIT4 phosphatase-associated family protein | Cre12.g549100.t3.1 U2 snRNP auxilliary factor, large subunit, splicing factor |
| Cre12.g559800.t1.2 microsomal glutathione s-transferase, putative | Cre12.g560300.t3.1 RNA-binding (RRM/RBD/RNP motifs) family protein |
| Cre13.g573400.t1.2 | Cre13.g577850.t1.1 FKBP-like peptidyl-prolyl cis-trans isomerase family protein |
| Cre13.g580000.t2.1 | Cre13.g602650.t1.2 Rhodanese/Cell cycle control phosphatase superfamily protein |
| Cre14.g615250.t1.1 Thioredoxin superfamily protein | Cre17.g733250.t1.2 FKBP-like peptidyl-prolyl cis-trans isomerase family protein |
|
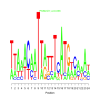
e.value: 0.058 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: TtAaaCaATTattTcATgtcaaAn |
motif_0133_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.000000039 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: CAGCaGCAGCAacAG |
Displaying 1 - 2 of 2
| Interaction | Weight | |
|---|---|---|
| down-regulates |
Cre13.g586450.t1.2Submitted by admin on Tue, 05/19/2015 - 15:51 |
0.068729 |
| up-regulates |
Cre06.g268600.t1.2Submitted by admin on Tue, 05/19/2015 - 15:51glycine-rich protein 2B |
0.035543 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

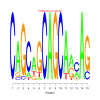
Comments