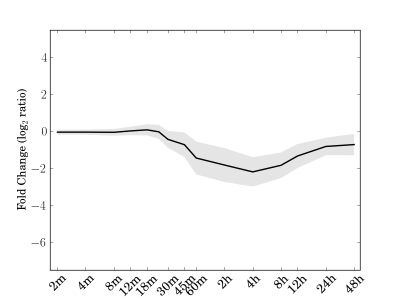
Module 169 Residual: 0.25
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0169 | v02 | 0.25 | -12.20 |
Displaying 1 - 20 of 20
| Cre01.g017300.t1.2 Plastid ribosomal protein S21 | Cre02.g076600.t1.2 Peptidyl-tRNA hydrolase family protein |
| Cre02.g077350.t1.2 histidinol dehydrogenase | Cre02.g109550.t1.2 aspartate kinase-homoserine dehydrogenase ii |
| Cre03.g161400.t1.2 tryptophan synthase beta-subunit 2 | Cre03.g189400.t1.2 seryl-tRNA synthetase / serine--tRNA ligase |
| Cre04.g220633.t1.1 | Cre05.g238322.t1.1 Nucleotidylyl transferase superfamily protein |
| Cre06.g250902.t1.1 Homocysteine S-methyltransferase family protein | Cre06.g308533.t2.1 |
| Cre07.g350500.t1.2 ATP binding;valine-tRNA ligases;aminoacyl-tRNA ligases;nucleotide binding;ATP binding;aminoacyl-tRNA ligases | Cre08.g375550.t1.2 |
| Cre09.g389450.t1.2 U-box domain-containing protein kinase family protein | Cre10.g433000.t1.1 glycine-tRNA ligases |
| Cre10.g436350.t1.1 shikimate kinase 1 | Cre12.g494750.t1.2 chloroplast 30S ribosomal protein S20, putative |
| Cre12.g511900.t1.2 D-ribulose-5-phosphate-3-epimerase | Cre12.g560550.t1.1 formyltransferase, putative |
| Cre16.g662150.t1.2 cofactor assembly of complex C | Cre17.g739550.t1.1 Domain of unknown function (DUF1995) |
|
e.value: 0.0003 Motif Bicluster: Width: 14 Number of Sites: 1 Consensus: ACACAcGCACaCgC |
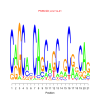
motif_0169_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 2.1e-21 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: CaGCacCAgCagCagCggCAg |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0043038 | GO:0043039 | GO:1901564 | GO:0006399 | GO:0016874 | amino acid activation | tRNA aminoacylation | organonitrogen compound metabolic proces... | tRNA metabolic process | ligase activity |

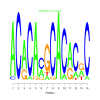
Comments