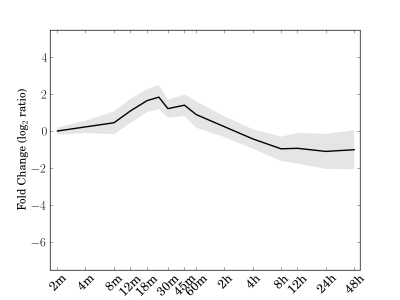
Module 192 Residual: 0.25
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0192 | v02 | 0.25 | -12.98 |
Displaying 1 - 24 of 24
| Cre01.g038950.t1.2 selenoprotein family protein | Cre02.g088000.t1.2 prohibitin 1 |
| Cre02.g106300.t1.2 Protein of unknown function (DUF1682) | Cre02.g118550.t1.2 |
| Cre03.g151600.t1.2 | Cre03.g151650.t1.1 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| Cre03.g180450.t1.2 5\'-nucleotidase and Flagellar Associated Protein | Cre03.g195200.t1.2 alpha/beta-Hydrolases superfamily protein |
| Cre06.g257650.t1.2 peptide met sulfoxide reductase 4 | Cre06.g295400.t1.2 Mitochondrial substrate carrier family protein |
| Cre06.g295450.t1.2 hydroxypyruvate reductase | Cre06.g304850.t1.1 Protein of unknown function (DUF544) |
| Cre06.g307500.t1.1 Low-CO2 inducible protein | Cre06.g309100.t1.2 heat shock protein 60 |
| Cre07.g324200.t1.2 Diacylglyceryl-N,N,N-trimethylhomoserine synthesis protein | Cre08.g360950.t1.1 |
| Cre10.g448950.t1.1 DNAse I-like superfamily protein | Cre10.g466000.t1.2 Cytochrome c oxidase, subunit Vib family protein |
| Cre12.g534800.t1.1 glycine decarboxylase P-protein 2 | Cre12.g542300.t1.2 P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| Cre16.g654400.t1.1 | Cre16.g657300.t1.2 cyclopropyl isomerase |
| Cre16.g669525.t1.1 non-intrinsic ABC protein 8 | Cre16.g691800.t1.1 |
|
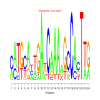
e.value: 0.0000058 Motif Bicluster: Width: 20 Number of Sites: 1 Consensus: CCgCCaCnGCCACaGCCgCC |
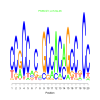
motif_0192_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.0097 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: cATGcAttGAgCAAAaGCCAcTTG |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

Comments