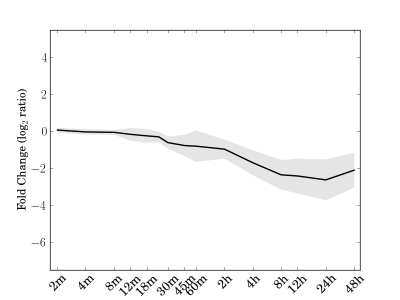
Module 80 Residual: 0.16
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0080 | v02 | 0.16 | -13.23 |
Displaying 1 - 19 of 19
| Cre01.g005950.t1.2 FAD/NAD(P)-binding oxidoreductase | Cre02.g076250.t1.1 Translation elongation factor EFG/EF2 protein |
| Cre02.g088850.t1.2 6,7-dimethyl-8-ribityllumazine synthase / DMRL synthase / lumazine synthase / riboflavin synthase | Cre03.g146567.t1.1 |
| Cre03.g158750.t1.1 P-loop containing nucleoside triphosphate hydrolases superfamily protein | Cre03.g170800.t1.2 |
| Cre03.g182150.t1.2 thylakoid lumen 18.3 kDa protein | Cre05.g243050.t1.2 thioredoxin F-type 1 |
| Cre06.g255500.t1.2 alpha/beta-Hydrolases superfamily protein | Cre06.g256250.t1.2 chloroplast thylakoid lumen protein |
| Cre06.g278104.t1.1 | Cre07.g331500.t1.1 ATP-dependent caseinolytic (Clp) protease/crotonase family protein |
| Cre07.g341600.t2.1 Co-chaperone GrpE family protein | Cre09.g404000.t1.1 Tetratricopeptide repeat (TPR)-like superfamily protein |
| Cre12.g510400.t1.1 rubredoxin family protein | Cre12.g543900.t1.1 |
| Cre13.g562900.t1.2 | Cre16.g665250.t1.2 acclimation of photosynthesis to environment |
| Cre16.g680450.t1.1 |
|
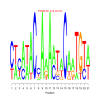
e.value: 140 Motif Bicluster: Width: 23 Number of Sites: 1 Consensus: GggGaAAtggATggtgAAGnAGA |
motif_0080_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 6300 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: CtcAtACcAaAaTaCAaTGTa |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0009349 | riboflavin synthase complex |

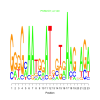
Comments