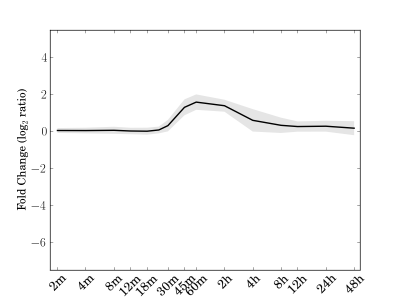
Module 113 Residual: 0.26
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0113 | v02 | 0.26 | -10.17 |
Displaying 1 - 30 of 30
| Cre01.g051625.t1.1 | Cre03.g144727.t1.1 RHOMBOID-like protein 14 |
| Cre03.g201327.t1.1 NAD(P)-linked oxidoreductase superfamily protein | Cre04.g217945.t2.1 fatty acid desaturase 5 |
| Cre05.g243455.t1.1 | Cre06.g260950.t1.2 EPS15 homology domain 1 |
| Cre06.g263289.t1.2 Undecaprenyl pyrophosphate synthetase family protein | Cre06.g278124.t1.1 Vacuolar sorting protein 39 |
| Cre06.g290800.t1.1 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Cre07.g320800.t1.2 |
| Cre07.g342000.t1.1 protein-l-isoaspartate methyltransferase 1 | Cre07.g357350.t1.2 GLNB1 homolog |
| Cre08.g358574.t1.1 | Cre09.g388393.t1.1 exostosin family protein |
| Cre09.g399050.t1.1 SGNH hydrolase-type esterase superfamily protein | Cre09.g401145.t1.1 |
| Cre10.g437000.t1.2 | Cre10.g441100.t1.2 GPI transamidase component Gpi16 subunit family protein |
| Cre10.g456400.t1.1 Dicarboxylate/amino acid cation sodium transporter | Cre10.g464133.t1.1 |
| Cre11.g467559.t1.1 Leucine-rich repeat family protein | Cre12.g490800.t1.1 Mitogen activated protein kinase kinase kinase-related |
| Cre12.g498750.t1.1 Myzus persicae-induced lipase 1 | Cre12.g508852.t1.1 |
| Cre12.g537750.t1.2 RabGAP/TBC domain-containing protein | Cre12.g542400.t1.1 damaged DNA binding;DNA-directed DNA polymerases |
| Cre13.g607550.t1.2 | Cre14.g616250.t1.2 Polyketide cyclase / dehydrase and lipid transport protein |
| Cre14.g628500.t1.1 NAD(P)-binding Rossmann-fold superfamily protein | Cre17.g711600.t1.2 |
|
e.value: 0.0000016 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: GnTtaTGTTGtTGtT |
motif_0113_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.0000000000001 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: cTGCTGcTaCTGCTG |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
| down-regulates |
Cre13.g573000.t1.2Submitted by admin on Tue, 05/19/2015 - 15:52SET domain-containing protein |
0.061551 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0006808 | GO:0019740 | GO:0030234 | regulation of nitrogen utilization | nitrogen utilization | enzyme regulator activity |

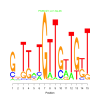
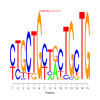
Comments