MSMEG_0766 MSMEG_0766 Phosphoribosylglycinamide formyltransferase 2 (EC 2.1.2.-)
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Locus Tag | Symbol | Synonyms | Product | Protein Synonyms | Start | End | Strand | PubMed ID | |
|---|---|---|---|---|---|---|---|---|---|
| MSMEG_0766 | MSMEG_0766 | purT | purT, MSMEG_0766 | Phosphoribosylglycinamide formyltransferase 2 (EC 2.1.2.-) |
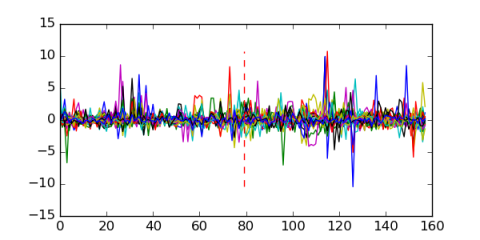
| Residual | Motif 1 | Expression Plot | |
|---|---|---|---|
| msm_bicluster_0198 | 0.54 |
 |
 |
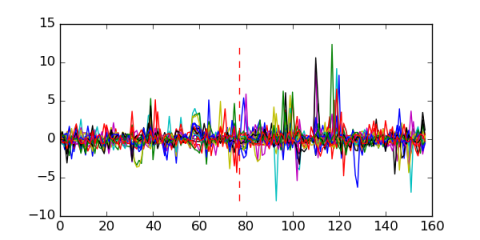
| msm_bicluster_0309 | 0.52 |
 |
 |
| Uniprot | NCBI Gene | NCBI Protein | EnsemblBacteria | InterPro | EggNOG | OrthoDB |
|---|---|---|---|---|---|---|
| A0QQI4 | 4532947; | WP_003892190.1, YP_885172.1 | ABK74865, AIU06030 | IPR011761, IPR003135, IPR013815, IPR013816, IPR016185, IPR011054 | COG0027, ENOG4107SPG | EOG6GJBVD |
| Title | Insert | TAG Module |
|---|---|---|
| NA |
| Orthologues | Paralogues |
|---|---|
| Catalytic Activity | KEGG Ontology | Pathways |
|---|---|---|
| Formate + ATP + 5'-phospho-ribosylglycinamide = 5'-phosphoribosyl-N-formylglycinamide + ADP + diphosphate, ECO:0000256|SAAS:SAAS00389763}. | msb:LJ00_03805, msm:MSMEG_0766 | Purine metabolism, IMP biosynthesis via de novo pathway, N(2)-formyl-N(1)-(5-phospho-D-ribosyl)glycinamide from N(1)-(5-phospho-D-ribosyl)glycinamide (formate route): step 1/1. {ECO:0000256|SAAS:SAAS00389753}. |
| Product (LegacyBRC) | Product (RefSeq) |
|---|---|
| GI Number | Accession | Blast | Conserved Domains |
|---|---|---|---|
| Link to STRINGS | STRINGS Network |
|---|---|
| MSMEG_0766 |
Add comment
Log in to post comments