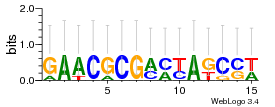
Module 222 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 0.0000000016 | 2100 | 79 | 32 | 20160204 |
| MSMEG_5227 Exodeoxyribonuclease 7 small subunit (EC 3.1.11.6) (Exodeoxyribonuclease VII small subunit) | MSMEG_4577 DegV family protein |
| MSMEG_5226 Exodeoxyribonuclease 7 large subunit (EC 3.1.11.6) | MSMEG_3124 FeS assembly ATPase SufC |
| MSMEG_4296 Protease (Tripeptidyl-peptidase B Serine peptidase MEROPS family S33) | MSMEG_3125 Cysteine desulfurase (EC 2.8.1.7) |
| MSMEG_4817 6-phosphogluconate dehydrogenase NAD-binding protein (EC 1.1.1.-) (6-phosphogluconate dehydrogenase, NAD-binding) | MSMEG_3126 SUF system FeS assembly protein (SUF system FeS assembly protein, NifU family protein) |
| MSMEG_4813 Conserved domain protein (Ferredoxin) | MSMEG_3127 Conserved protein, DUF59 (Putative metal-sulfur cluster biosynthetic enzyme) |
| MSMEG_4812 Respiratory-chain NADH dehydrogenase domain, 51 kDa subunit (EC 1.6.99.5) | MSMEG_3121 DNA-binding protein |
| MSMEG_4581 Probable nicotinate-nucleotide adenylyltransferase (EC 2.7.7.18) (Deamido-NAD(+) diphosphorylase) (Deamido-NAD(+) pyrophosphorylase) (Nicotinate mononucleotide adenylyltransferase) (NaMN adenylyltransferase) | MSMEG_3122 FeS assembly protein SufB |
| MSMEG_4580 Ribosomal silencing factor RsfS | MSMEG_3123 FeS assembly protein SufD |
| MSMEG_5791 Uncharacterized protein | MSMEG_5787 Uncharacterized protein |
| MSMEG_5793 Uncharacterized protein | MSMEG_2931 Threonine--tRNA ligase (EC 6.1.1.3) (Threonyl-tRNA synthetase) (ThrRS) |
| MSMEG_5792 UPF0678 fatty acid-binding protein-like protein MSMEG_5792/MSMEI_5639 | MSMEG_2932 HIT family protein hydrolase (Histidine triad (HIT) protein) |
| MSMEG_4187 Uncharacterized protein | MSMEG_2933 Phosphatidylinositol synthase (Putative pi synthase Pgsa1 (Phosphatidylinositol synthase) (Cdp-diacylglycerol--inositol3-phosphatidyltransferase)) (EC 2.7.8.-) |
| MSMEG_5031 Uracil-DNA glycosylase superfamily protein | MSMEG_2934 Phosphatidylinositol mannoside acyltransferase (PIM acyltransferase) (EC 2.3.1.-) |
| MSMEG_4188 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.243) | MSMEG_2935 GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase (EC 2.4.1.57) (Alpha-mannosyltransferase) (Guanosine diphosphomannose-phosphatidyl-inositol alpha-mannosyltransferase) (Phosphatidylinositol alpha-mannosyltransferase) (PI alpha-mannosyltransferase) |
| MSMEG_4578 Uncharacterized protein | MSMEG_2936 Hydrolase, nudix family protein (NUDIX hydrolase) |
| MSMEG_4579 Phosphoglycerate mutase (EC 5.4.2.-) (Phosphoglycerate mutase family protein) | MSMEG_5029 Alkanal monooxygenase alpha chain (Luciferase-like monooxygenase) |
KO_stationary.phase_rep1
Biological.replicate.5.of.7..falcarinol.treatment
Biological.replicate.2.of.7..panaxydol.treatment
dcpA.knockout.rep2
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
dosR.WT_Replicate2
dosR.WT_Replicate1
Biological.replicate.2.of.6..kanamycin.treatment
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Wild.type.rep3
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.4.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.6..negative.control
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.5.of.7..panaxydol.treatment
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide601norm
X2.5..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..ethambutol.treatment
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X4XR.rep2
X4XR.rep1
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.with.vs..without.N_rep2
WT.vs..DglnR.N_starvation_rep2
Wild.type.rep2.x
Biological.replicate.1.of.6..negative.control
X2XR.rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DglnR.N_starvation_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.3.of.6..negative.control
WT.vs..DamtR.N.surplus_rep2
Comments
Log in to post comments