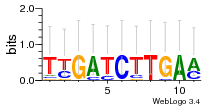
Module 505 Residual: 0.57
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.57 | 0.00000000000094 | 0.00000000013 | 74 | 27 | 20160204 |
| MSMEG_2270 Uncharacterized protein | MSMEG_6665 Integral membrane protein (Uncharacterized protein) |
| MSMEG_1109 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase (SEPHCHC synthase) (EC 2.2.1.9) (Menaquinone biosynthesis protein MenD) | MSMEG_5435 Putative ligase MSMEG_5435/MSMEI_5285 (EC 6.2.1.-) |
| MSMEG_0069 Translation initiation factor IF-2 protein | MSMEG_4703 Glycerol-3-phosphate acyltransferase (Putative acyltransferase plsB1) (EC 2.3.1.15) |
| MSMEG_5970 Glutamate-1-semialdehyde 2,1-aminomutase (EC 5.4.3.8) (Probable glutamate-1-semialdehyde 2,1-aminomutase) (EC 5.4.3.8) | MSMEG_4705 Diacylglycerol O-acyltransferase (EC 2.3.1.20) |
| MSMEG_0218 3-demethylubiquinone-9 3-methyltransferase | MSMEG_4704 Acyltransferase family protein |
| MSMEG_6314 Haloalkane dehalogenase (Haloalkane dehalogenase 1) (EC 3.8.1.5) | MSMEG_6329 Uncharacterized protein |
| MSMEG_6316 Lipoprotein LpqH (Lipoprotein lpqH) | MSMEG_5848 Amino acid permease-associated region |
| MSMEG_5851 Conserved membrane protein (Putative esterase superfamily protein) | MSMEG_0671 Alcohol dehydrogenase GroES domain protein (S-(Hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) |
| MSMEG_4973 Glyoxalase/bleomycin resistance protein/dioxygenase | MSMEG_2269 NHL repeat protein |
| MSMEG_3304 Succinate semialdehyde dehydrogenase | MSMEG_5969 Uncharacterized protein |
| MSMEG_3329 Uncharacterized protein | MSMEG_0217 Alcohol dehydrogenase B (EC 1.1.1.1) (Zinc-type alcohol dehydrogenase AdhD) |
| MSMEG_3412 Polysaccharide deacetylase (Polysaccharide deacetylase family protein) | MSMEG_1232 ABC transporter substrate-binding protein (Nitrate ABC transporter, periplasmic nitrate-binding protein, putative (Putative ABC-type nitrate/sulfonate/bicarbonate transport system periplasmic component)) |
| MSMEG_3276 Integral membrane protein (Putative integral membrane protein (DMT superfamily)) | MSMEG_1231 Inner membrane protein YidH |
| MSMEG_4618 Isochorismatase family protein (Isochorismatase hydrolase) |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
dcpA.knockout.rep2
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Biological.replicate.4.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.30min
dosR.WT_Replicate3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
Wild.type.rep2.y
dosR.WT_Replicate4
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.2
dosR.WT_Replicate2
X0.6..vs..50..air.saturation.Replicate.4
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Plogcy5plogcy3slide111norm
X2.5..vs..50..air.saturation.Replicate.4
stphcy3plogcy5slide581norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.2
WT_stationary.phase_rep2
WT_stationary.phase_rep1
X4XR.rep2
X4XR.rep1
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_dyeswap_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
Biological.replicate.2.of.6..negative.control
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..kanamycin.treatment
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments