MSMEG_5832 MSMEG_5832 Phosphoribosylformylglycinamidine synthase subunit PurS (FGAM synthase) (EC 6.3.5.3) (Formylglycinamide ribonucleotide amidotransferase subunit III) (Phosphoribosylformylglycinamidine synthase subunit III)
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Locus Tag | Symbol | Synonyms | Product | Protein Synonyms | Start | End | Strand | PubMed ID | |
|---|---|---|---|---|---|---|---|---|---|
| MSMEG_5832 | MSMEG_5832 | purS | purS, MSMEG_5832 | Phosphoribosylformylglycinamidine synthase subunit PurS (FGAM synthase) (EC 6.3.5.3) (Formylglycinamide ribonucleotide amidotransferase subunit III) (Phosphoribosylformylglycinamidine synthase subunit III) |
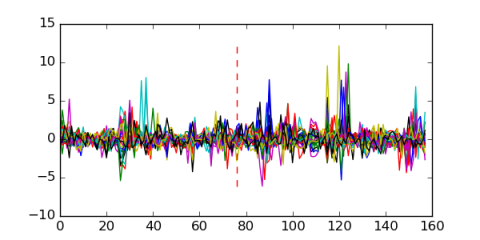
| Residual | Motif 1 | Expression Plot | |
|---|---|---|---|
| msm_bicluster_0352 | 0.55 |
 |
 |
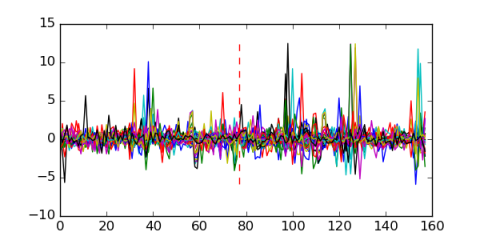
| msm_bicluster_0627 | 0.52 |
 |
 |
| Uniprot | NCBI Gene | NCBI Protein | EnsemblBacteria | InterPro | EggNOG | OrthoDB |
|---|---|---|---|---|---|---|
| A0R4H2 | 4531281; | WP_003897235.1, YP_890060.1 | ABK75985 | IPR003850 | COG1828, ENOG410839H | EOG6TFCXJ |
| Title | Insert | TAG Module |
|---|---|---|
| NA |
| Orthologues | Paralogues |
|---|---|
| Catalytic Activity | KEGG Ontology | Pathways |
|---|---|---|
| ATP + N(2)-formyl-N(1)-(5-phospho-D-ribosyl)glycinamide + L-glutamine + H(2)O = ADP + phosphate + 2-(formamido)-N(1)-(5-phospho-D-ribosyl)acetamidine + L-glutamate, ECO:0000256|HAMAP-Rule:MF_01926}. | msm:MSMEG_5832 | Purine metabolism, IMP biosynthesis via de novo pathway, 5-amino-1-(5-phospho-D-ribosyl)imidazole from N(2)-formyl-N(1)-(5-phospho-D-ribosyl)glycinamide: step 1/2. {ECO:0000256|HAMAP-Rule:MF_01926}. |
| Product (LegacyBRC) | Product (RefSeq) |
|---|---|
| GI Number | Accession | Blast | Conserved Domains |
|---|---|---|---|
| Link to STRINGS | STRINGS Network |
|---|---|
| MSMEG_5832 |
Add comment
Log in to post comments