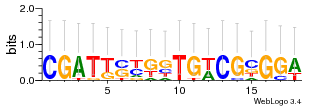
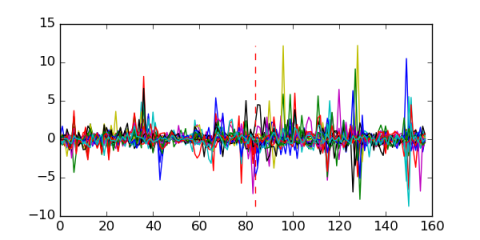
Module 74 Residual: 0.50
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 0.000026 | 0.00000057 | 84 | 25 | 20160204 |
| MSMEG_5049 Multifunctional 2-oxoglutarate metabolism enzyme (2-hydroxy-3-oxoadipate synthase) (HOA synthase) (HOAS) (EC 2.2.1.5) (2-oxoglutarate carboxy-lyase) (2-oxoglutarate decarboxylase) (Alpha-ketoglutarate decarboxylase) (KG decarboxylase) (KGD) (EC 4.1.1.71) (Alpha-ketoglutarate-glyoxylate carboligase) [Includes: 2-oxoglutarate dehydrogenase E1 component (ODH E1 component) (EC 1.2.4.2) (Alpha-ketoglutarate dehydrogenase E1 component) (KDH E1 component); Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex (EC 2.3.1.61) (2-oxoglutarate dehydrogenase complex E2 component) (ODH E2 component) (OGDC-E2) (Dihydrolipoamide succinyltransferase)] | MSMEG_1830 2-phospho-L-lactate transferase (EC 2.7.8.28) (LPPG:FO 2-phospho-L-lactate transferase) |
| MSMEG_0264 Transmembrane transport protein | MSMEG_0403 Peptidoglycolipid exporter Gap (GPLs addressing protein) |
| MSMEG_1827 Hydrolase, nudix family protein, putative (NUDIX hydrolase) | MSMEG_0401 Putative non-ribosomal peptide synthase |
| MSMEG_1829 Coenzyme F420:L-glutamate ligase (EC 6.3.2.31) (EC 6.3.2.34) (Coenzyme F420-0:L-glutamate ligase) (Coenzyme F420-1:gamma-L-glutamate ligase) | MSMEG_5009 ABC transporter |
| MSMEG_0402 Linear gramicidin synthetase subunit D (EC 1.-.-.-) (EC 5.1.1.-) (Non-ribosomal peptide synthetase) | MSMEG_5008 ABC transporter, ATP-binding protein (Transmembrane ATP-binding protein ABC transporter) |
| MSMEG_5797 Uncharacterized protein | MSMEG_0040 NIPSNAP domain containing protein (Uncharacterized protein) |
| MSMEG_1831 Transcriptional regulator WhiB2 | MSMEG_0041 Uncharacterized protein |
| MSMEG_0039 Small membrane hydrophobic protein (Uncharacterized protein) | MSMEG_5005 Lipoprotein LprC (LprC protein) |
| MSMEG_4898 Lipoprotein lprD (Uncharacterized protein) | MSMEG_0003 DNA replication and repair protein RecF |
| MSMEG_3731 Dipeptidase (EC 3.4.13.19) (Dipeptidase 2) | MSMEG_5007 LprB protein |
| MSMEG_4205 Putative transcriptional regulator | MSMEG_0001 DNA polymerase III subunit beta (EC 2.7.7.7) |
| MSMEG_0002 6-phosphogluconate dehydrogenase (EC 1.1.1.44) (6-phosphogluconate dehydrogenase, decarboxylating) | MSMEG_0004 UPF0232 protein MSMEG_0004/MSMEI_0006 |
| MSMEG_4900 Conserved membrane protein (Pks14 protein) |
Wild.type.rep2.x
WT.vs..DglnR.N_starvation_rep2
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.4.of.6..negative.control
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
Plogcy5plogcy3slide110norm
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
Biological.replicate.5.of.6..kanamycin.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
KO_log.phase_rep2
KO_log.phase_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Wild.type.rep3
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.6..negative.control
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
X0.6..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DamtR.N.surplus_rep1
dosR.WT_Replicate4
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
X3dbfcy5plogcy3slide603norm
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
WT_log.phase_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
WT.with.vs..without.N_dye.swap_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
WT.vs..DglnR.N_starvation_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..kanamycin.treatment
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments