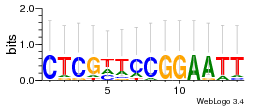
Module 353 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 5.6e-17 | 0.028 | 75 | 31 | 20160204 |
| MSMEG_3708 Catalase-related peroxidase (EC 1.11.1.-) | MSMEG_6848 Putative oxidoreductase (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_0821 Glutamyl-tRNA(Gln) amidotransferase subunit A (EC 6.3.5.-) | MSMEG_6602 Oxidoreductase (Putative luciferase-like oxidoreductase) |
| MSMEG_4917 Tetratricopeptide repeat domain protein (Thioredoxin) | MSMEG_0850 Uncharacterized protein |
| MSMEG_6541 Anti-sigma factor antagonist | MSMEG_3436 Uncharacterized protein |
| MSMEG_4919 Uncharacterized protein | MSMEG_3022 Putative transglycosylase associated protein (Transglycosylase associated protein) |
| MSMEG_6542 B12 binding domain protein (Cobalamin B12-binding protein) | MSMEG_2549 Major facilitator superfamily MFS_1 (Major facilitator superfamily protein) |
| MSMEG_2550 D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding protein (Glyoxylate reductase) (EC 1.1.1.26) | MSMEG_4414 Uncharacterized protein |
| MSMEG_0705 Major facilitator superfamily MFS_1 (Putative permease of the major facilitator superfamily protein) | MSMEG_0819 Amidase, hydantoinase/carbamoylase (EC 3.5.1.87) (N-carbamoyl-L-amino acid amidohydrolase) |
| MSMEG_4483 Deoxyguanosinetriphosphate triphosphohydrolase-like protein | MSMEG_0895 Bacterial regulatory protein, GntR family protein (GntR family transcriptional regulator) |
| MSMEG_4484 Putative conserved transmembrane protein | MSMEG_2286 |
| MSMEG_3967 TonB-dependent receptor | MSMEG_0047 Uncharacterized protein |
| MSMEG_1088 Asp-tRNAAsn/Glu-tRNAGln amidotransferase A subunit-like amidase (EC 3.5.1.4) (Glutamyl-tRNA(Gln)/aspartyl-tRNA(Asn) amidotransferase, A subunit) (EC 6.3.5.-) | MSMEG_3838 Integral membrane protein |
| MSMEG_3710 Cytochrome B561 (Cytochrome b561 family protein) | MSMEG_3836 Uncharacterized protein |
| MSMEG_1269 Diadenosine tetraphosphatase and related serine/threonine protein phosphatase (EC 3.6.1.41) (Ser/Thr protein phosphatase family protein) | MSMEG_3834 Putative TetR-family protein transcriptional regulator (Transcriptional regulator, TetR family) |
| MSMEG_6749 Uncharacterized protein | MSMEG_3835 Uncharacterized protein (YesF protein) |
| MSMEG_5748 Uncharacterized protein |
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate1
Biological.replicate.2.of.7..panaxydol.treatment
Plogcy5plogcy3slide110norm
Wild.type.rep1.y
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
Biological.replicate.3.of.7..ethambutol.treatment
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Slow.vs..Fast.Replicate.3
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.4
Biological.replicate.4.of.7..ethambutol.treatment
stphcy3plogcy5slide581norm
WT_log.phase_rep1
DglnR_without.vs..with.N_rep1
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
WT_stationary.phase_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X4XR.rep2
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
dosR.WT_Replicate3
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DamtR.N.surplus_dyeswap_rep2
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X4dbfcy5plogcy3slide480norm
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep1
X2XR.rep1
Biological.replicate.2.of.6..negative.control
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
Biological.replicate.2.of.7..falcarinol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.6.of.7..falcarinol.treatment
Comments
Log in to post comments