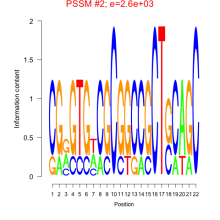
Phatr_bicluster_0148 Residual: 0.35
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0148 | 0.35 | Phaeodactylum tricornutum |
Displaying 1 - 33 of 33
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10024 heterogeneous nuclear ribonucleoprotein h3 (RRM_hnRNPH_ESRPs_RBM12_like)
PHATRDRAFT_11259 epc1 (ectopically parting cells)transferring glycosyl groups (Glyco_transf_64 superfamily)
PHATRDRAFT_11826 sugar transporter family protein (SP)
PHATRDRAFT_14699 novel proteinvertebrate ring finger and chy zinc finger domain containing 1(zgc:101128) (zf-CHY superfamily)
PHATRDRAFT_14770 cytochrome c oxidase assembly factor (SCO)
PHATRDRAFT_18469 d-xylose proton-symporter (Sugar_tr)
PHATRDRAFT_2808 protein disulfide isomerase associated 6 (Thioredoxin_like superfamily)
PHATRDRAFT_31684 soul heme-binding protein (DUF2358 superfamily)
PHATRDRAFT_32813 glutathione s-transferase-like protein
PHATRDRAFT_33590 major facilitator family transporter (2A0106)
PHATRDRAFT_35054 agc family protein kinase (S_TKc)
PHATRDRAFT_35296 loc100125051 protein (MATE_like superfamily)
PHATRDRAFT_36109 (MMACHC-like)
PHATRDRAFT_41069 (PAP_fibrillin superfamily)
PHATRDRAFT_43301 integral membrane protein (Ureide_permease superfamily)
PHATRDRAFT_45216 (PqiA superfamily)
PHATRDRAFT_45504 dehydrogenase reductase 1 (NADB_Rossmann superfamily)
PHATRDRAFT_46136 phosphatidate cytidylyltransferase (ADP_ribosyl_GH)
PHATRDRAFT_46389 aspartyl protease family protein (pepsin_retropepsin_like superfamily)
PHATRDRAFT_47564 endomembrane protein (EMP70 superfamily)
PHATRDRAFT_48170 3-phosphoinositide-dependent protein kinase 1 (S_TKc)
PHATRDRAFT_48882 light-harvesting chlorophyll a-c binding protein (Chloroa_b-bind superfamily)
PHATRDRAFT_49462 diacylglycerol (LPLAT_MGAT-like)
PHATRDRAFT_8154 fk506 binding protein 7 (FKBP_C)
PHATRDRAFT_83 abc transporter (ABCG_PDR_domain2)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008643 | carbohydrate transport | 0.00192368 | 1 | Phatr_bicluster_0148 |
| GO:0006118 | electron transport | 0.249468 | 1 | Phatr_bicluster_0148 |
| GO:0008152 | metabolism | 0.723107 | 1 | Phatr_bicluster_0148 |
| GO:0008654 | phospholipid biosynthesis | 0.0371948 | 1 | Phatr_bicluster_0148 |
| GO:0009765 | photosynthesis light harvesting | 0.151992 | 1 | Phatr_bicluster_0148 |
| GO:0006468 | protein amino acid phosphorylation | 0.161086 | 1 | Phatr_bicluster_0148 |
| GO:0006457 | protein folding | 0.393149 | 1 | Phatr_bicluster_0148 |
| GO:0016567 | protein ubiquitination | 0.263338 | 1 | Phatr_bicluster_0148 |
| GO:0006508 | proteolysis and peptidolysis | 0.620715 | 1 | Phatr_bicluster_0148 |
| GO:0006979 | response to oxidative stress | 0.0963076 | 1 | Phatr_bicluster_0148 |


Comments