Phatr_bicluster_0113 Residual: 0.23
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0113 | 0.23 | Phaeodactylum tricornutum |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12188 emb1241 (embryo defective 1241) adenyl-nucleotide exchange factor chaperone binding protein binding protein homodimerization (GrpE)
PHATRDRAFT_12572 der1-like domainmember 3 (DER1 superfamily)
PHATRDRAFT_3061 c-terminal processing peptidase (Peptidase_S41_CPP)
PHATRDRAFT_32554 magnesium-dependent phosphatase 1 (Acid_PPase)
PHATRDRAFT_36597 transitional endoplasmic reticulum atpase ter94 (CDC48)
PHATRDRAFT_41850 cell division cycle proteinexpressed (SpoVK)
PHATRDRAFT_42426 prolyl 4-hydroxylase alpha subunit (2OG-FeII_Oxy_3 superfamily)
PHATRDRAFT_42903 thioredoxin
PHATRDRAFT_42980 phytoene dehydrogenase and related protein (COG1233)
PHATRDRAFT_44315 protein kinasereceptor type precursor
PHATRDRAFT_44344 inositol polyphosphate-5-phosphatase inp51p (EEP superfamily)
PHATRDRAFT_44790 phosphatidylinositol glycan anchorclass b (Glyco_transf_22 superfamily)
PHATRDRAFT_54068 glycosylgroup 1 family protein (GT1_PIG-A_like)
PHATRDRAFT_54197 chloroplast envelope protein translocase (cept or tic-toc) family protein (Tic22 superfamily)
PHATRDRAFT_6554 (FMN_red superfamily)
PHATRDRAFT_7525 atp-dependent clp protease adaptor proteincontaining protein (ClpS)
PHATRDRAFT_9956 ac008051_10identical to gene zw10 from arabidopsis thaliana gb (His_Phos_1)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019538 | protein metabolism | 0.0264433 | 1 | Phatr_bicluster_0113 |
| GO:0006457 | protein folding | 0.0842275 | 1 | Phatr_bicluster_0113 |
| GO:0006508 | proteolysis and peptidolysis | 0.156991 | 1 | Phatr_bicluster_0113 |

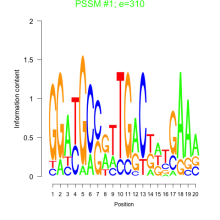

Comments