DVU1425 DVU1425 Probable glycine dehydrogenase
| Locus Tag | Symbol | Synonyms | Product | Protein Synonyms | Start | End | Strand | PubMed ID | |
|---|---|---|---|---|---|---|---|---|---|
| DVU1425 | DVU1425 | gcvPA | gcvPA, DVU1425 | Probable glycine dehydrogenase | Probable glycine dehydrogenase (decarboxylating) subunit 1 (EC 1.4.4.2) (Glycine cleavage system P-protein subunit 1) (Glycine decarboxylase subunit 1) (Glycine dehydrogenase (aminomethyl-transferring) subunit 1) | 1500690 | 1499359 | - | 15077118 |
| Residual | Motif 1 | Expression Plot | |
|---|---|---|---|
| bicluster_0014 | 0.50 |
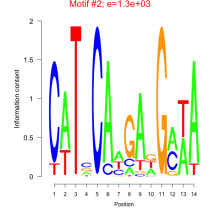 |
| Line | Position | Effect | Impact | Class | Percent Frequency | Sample | Title |
|---|---|---|---|---|---|---|---|
| UE3 | 1 499 475 | NON_SYNONYMOUS_CODING | MISSENSE | 98 | UE3.152.09.D01 | Chr-DVU1425-1499475-UE3.152.09.D01 |
| Title | Insert | TAG Module |
|---|---|---|
| GZ10775 | 96 | TagModule_3741 |
| GZ4541 | 241 | TagModule_0996 |
| GZ8285 | 269 | TagModule_1737 |
| GZ2683 | 278 | TagModule_1059 |
| GZ2706 | 300 | TagModule_1103 |
| GZ12603 | 734 | TagModule_3538 |
| GZ2943 | 822 | TagModule_0024 |
| GZ4825 | 822 | TagModule_0603 |
| GZ2146 | 846 | TagModule_0232 |
| GZ1636 | 1175 | TagModule_0747 |
| Orthologues | Paralogues |
|---|---|
| Catalytic Activity | COG | EC Description | Protein families |
|---|---|---|---|
| Glycine + [glycine-cleavage complex H protein]-N(6)-lipoyl-L-lysine = [glycine-cleavage complex H protein]-S-aminomethyl-N(6)-dihydrolipoyl-L-lysine + CO(2). {ECO:0000255|HAMAP-Rule:MF_00712}. | (E) COG403 | Glycine cleavage system protein P (pyridoxal-binding), N-terminal domain | (1.4.4.2) Glycine dehydrogenase (decarboxylating). | GcvP family, N-terminal subunit subfamily |
| Product (LegacyBRC) | Product (RefSeq) |
|---|---|
| GI Number | Accession | Blast | Conserved Domains |
|---|---|---|---|
| 46579836 | YP_010644.1 | Run |
| Link to STRINGS | STRINGS Network |
|---|---|
| DVU1425 |

Add comment