Methanococcus maripaludis S2 Module 84 Residual: 0.11
| Residual: 0.11 Score: -24.5681 version: mmp_cmonkey_11_Oct_11 |
| MMP1160 conserved hypothetical protein (NCBI) | MMP1156 conserved hypothetical protein (NCBI) |
| MMP0913 conserved hypothetical archaeal protein (NCBI) | MMP1157 Desulfoferrodoxin, ferrous iron-binding site (NCBI) |
| MMP0914 conserved hypothetical archaeal protein (NCBI) | MMP1158 conserved hypothetical protein (NCBI) |
| MMP1154 heterosulfide reductase, subunit C1 (NCBI) | MMP1159 Ferritin (NCBI) |
| MMP1155 heterosulfide reductase, subunit B1 (NCBI) | MMP1161 Conserved hypothetical Archael protein (NCBI) |
| Category | GO ID | GO Term | Annotated | Expected | Significant | p-value |
|---|---|---|---|---|---|---|
| BP | GO:0006873 | cellular ion homeostasis | 3 | 0.02 | 1 | 0.0160 |
| BP | GO:0048878 | chemical homeostasis | 3 | 0.02 | 1 | 0.0160 |
| BP | GO:0050801 | ion homeostasis | 3 | 0.02 | 1 | 0.0160 |
| BP | GO:0055082 | cellular chemical homeostasis | 3 | 0.02 | 1 | 0.0160 |
| BP | GO:0098771 | inorganic ion homeostasis | 3 | 0.02 | 1 | 0.0160 |
| BP | GO:0019725 | cellular homeostasis | 7 | 0.04 | 1 | 0.0360 |
| BP | GO:0042592 | homeostatic process | 7 | 0.04 | 1 | 0.0360 |
| BP | GO:0065008 | regulation of biological quality | 7 | 0.04 | 1 | 0.0360 |
| Category | GO ID | GO Term | Annotated | Expected | Significant | p-value |
|---|---|---|---|---|---|---|
| MF | GO:0005506 | iron ion binding | 4 | 0.03 | 2 | 0.0002 |
| MF | GO:0046914 | transition metal ion binding | 9 | 0.06 | 2 | 0.0012 |
| MF | GO:0046872 | metal ion binding | 24 | 0.16 | 2 | 0.0087 |
| MF | GO:0043169 | cation binding | 26 | 0.17 | 2 | 0.0102 |
| MF | GO:0016209 | antioxidant activity | 3 | 0.02 | 1 | 0.0193 |
| MF | GO:0016667 | oxidoreductase activity, acting on a sul... | 3 | 0.02 | 1 | 0.0193 |
| MF | GO:0016684 | oxidoreductase activity, acting on perox... | 3 | 0.02 | 1 | 0.0193 |











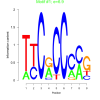

Comments