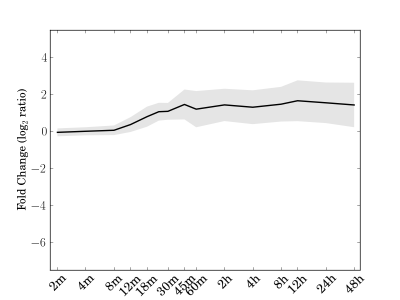
|
Cre01.g012750.t1.1 gated outwardly-rectifying K+ channel
|
Cre01.g044550.t1.2
Details for Cre01.g044550.t1.2 -
GO Terms: NA
Go to Gene Page: Cre01.g044550.t1.2
Phytozome Genomic Region

|
Cre01.g046800.t1.1
Details for Cre01.g046800.t1.1 -
GO Terms: NA
Go to Gene Page: Cre01.g046800.t1.1
Phytozome Genomic Region

|
Cre01.g053000.t1.2 NAD-dependent glycerol-3-phosphate dehydrogenase family protein
|
|
Cre02.g113100.t1.2 Uncharacterised protein family (UPF0041)
|
Cre03.g156476.t1.1
Details for Cre03.g156476.t1.1 -
GO Terms: NA
Go to Gene Page: Cre03.g156476.t1.1
Phytozome Genomic Region

|
Cre03.g199199.t1.1
Details for Cre03.g199199.t1.1 -
GO Terms: NA
Go to Gene Page: Cre03.g199199.t1.1
Phytozome Genomic Region

|
Cre03.g200400.t1.2
|
|
Cre03.g207650.t1.2
|
Cre04.g217350.t1.2 high affinity K+ transporter 5
|
Cre05.g245500.t1.1 Flagellar Associated Protein with ankyrin repeats
Details for Cre05.g245500.t1.1 - Flagellar Associated Protein with ankyrin repeats
GO Terms: GO:0005515
Go to Gene Page: Cre05.g245500.t1.1
Phytozome Genomic Region

|
Cre06.g282000.t2.1 starch synthase 3
|
Cre06.g288800.t1.2 Haloacid dehalogenase-like hydrolase (HAD) superfamily protein
Details for Cre06.g288800.t1.2 - Haloacid dehalogenase-like hydrolase (HAD) superfamily protein
GO Terms: NA
Go to Gene Page: Cre06.g288800.t1.2
Phytozome Genomic Region

|
Cre06.g310900.t1.2
Details for Cre06.g310900.t1.2 -
GO Terms: NA
Go to Gene Page: Cre06.g310900.t1.2
Phytozome Genomic Region

|
Cre07.g312850.t1.2 Protein of unknown function (DUF679)
Details for Cre07.g312850.t1.2 - Protein of unknown function (DUF679)
GO Terms: NA
Go to Gene Page: Cre07.g312850.t1.2
Phytozome Genomic Region

|
Cre07.g355050.t1.2
Details for Cre07.g355050.t1.2 -
GO Terms: NA
Go to Gene Page: Cre07.g355050.t1.2
Phytozome Genomic Region

|
Cre08.g361350.t1.2
Details for Cre08.g361350.t1.2 -
GO Terms: NA
Go to Gene Page: Cre08.g361350.t1.2
Phytozome Genomic Region

|
Cre08.g369350.t1.1
Details for Cre08.g369350.t1.1 -
GO Terms: NA
Go to Gene Page: Cre08.g369350.t1.1
Phytozome Genomic Region

|
|
Cre10.g423550.t1.1 threonine aldolase 2
|
Cre10.g456100.t1.2 flavodoxin-like quinone reductase 1
Details for Cre10.g456100.t1.2 - flavodoxin-like quinone reductase 1
GO Terms: GO:0016491
Go to Gene Page: Cre10.g456100.t1.2
Phytozome Genomic Region

|
Cre10.g456750.t1.2 dehydroascorbate reductase 2
Details for Cre10.g456750.t1.2 - dehydroascorbate reductase 2
GO Terms: NA
Go to Gene Page: Cre10.g456750.t1.2
Phytozome Genomic Region

|
Cre11.g467628.t1.1
Details for Cre11.g467628.t1.1 -
GO Terms: NA
Go to Gene Page: Cre11.g467628.t1.1
Phytozome Genomic Region

|
Cre12.g485478.t1.1
Details for Cre12.g485478.t1.1 -
GO Terms: NA
Go to Gene Page: Cre12.g485478.t1.1
Phytozome Genomic Region

|
Cre12.g507750.t2.1 Integrin-linked protein kinase family
|
|
Cre12.g510200.t1.2 Basic-leucine zipper (bZIP) transcription factor family protein
|
Cre12.g527918.t1.2
Details for Cre12.g527918.t1.2 -
GO Terms: NA
Go to Gene Page: Cre12.g527918.t1.2
Phytozome Genomic Region

|
|
Cre12.g537400.t1.1 ataurora3
|
Cre12.g543000.t1.1
Details for Cre12.g543000.t1.1 -
GO Terms: NA
Go to Gene Page: Cre12.g543000.t1.1
Phytozome Genomic Region

|
|
Cre12.g554850.t1.2 thioredoxin H-type 1
|
Cre13.g583450.t1.2 PLAC8 family protein
Details for Cre13.g583450.t1.2 - PLAC8 family protein
GO Terms: NA
Go to Gene Page: Cre13.g583450.t1.2
Phytozome Genomic Region

|
Cre16.g647950.t1.1
Details for Cre16.g647950.t1.1 -
GO Terms: NA
Go to Gene Page: Cre16.g647950.t1.1
Phytozome Genomic Region

|
Cre16.g673281.t1.1 phytanoyl-CoA dioxygenase (PhyH) family protein
Details for Cre16.g673281.t1.1 - phytanoyl-CoA dioxygenase (PhyH) family protein
GO Terms: NA
Go to Gene Page: Cre16.g673281.t1.1
Phytozome Genomic Region

|
|
Cre17.g726750.t1.2 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase 1
|
|

Comments