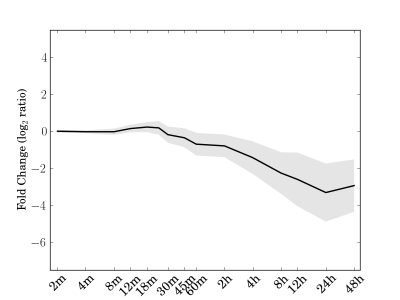
Module 185 Residual: 0.13
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0185 | v02 | 0.13 | -17.50 |
Displaying 1 - 19 of 19
| Cre01.g031100.t1.2 protein containing PDZ domain, a K-box domain, and a TPR region | Cre01.g033550.t1.2 thioredoxin family protein |
| Cre01.g043350.t1.2 Pheophorbide a oxygenase family protein with Rieske [2Fe-2S] domain | Cre01.g051500.t1.2 Uncharacterized thykaloid lumenal polypeptide |
| Cre03.g204601.t1.1 adenosine kinase 2 | Cre05.g236039.t1.2 |
| Cre06.g253350.t1.2 Single hybrid motif superfamily protein | Cre06.g259900.t1.2 ATPase, F1 complex, gamma subunit protein |
| Cre06.g261000.t1.2 photosystem II subunit R | Cre06.g263450.t1.2 GTP binding Elongation factor Tu family protein |
| Cre06.g278750.t1.2 plant uncoupling mitochondrial protein 1 | Cre06.g284600.t1.2 Rubredoxin-like superfamily protein |
| Cre08.g372450.t1.2 photosystem II subunit Q-2 | Cre08.g376000.t1.1 Rab5-interacting family protein |
| Cre09.g396213.t1.1 PS II oxygen-evolving complex 1 | Cre12.g544150.t1.2 cyclophilin 20-2 |
| Cre12.g550850.t1.2 photosystem II subunit P-1 | Cre17.g718468.t1.2 |
| Cre17.g733250.t1.2 FKBP-like peptidyl-prolyl cis-trans isomerase family protein |
|
e.value: 0.00000027 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: GCGGCGGCggCagcGGCgGcg |
motif_0185_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.048 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: atTGCATtGgGntCgagttacTTg |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
| down-regulates |
Cre12.g523000.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51zinc finger protein 1 |
0.065961 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0043234 | GO:0019898 | GO:0005509 | protein complex | extrinsic component of membrane | calcium ion binding |

Comments