35129 (Tp_Myb2R9) PLN03142Thalassiosira pseudonana
| Chromosome | Product | Transcript Start | End | Strand | Short Name | |
|---|---|---|---|---|---|---|
| 35129 | chr_6 | (Tp_Myb2R9) PLN03142 | 1631429 | 1636389 | + | PLN03142 |
| NCBI ID | Ensembl Genomes exon ID |
|---|---|
| 7442773 | Thaps35129.2, Thaps35129.3, Thaps35129.5, Thaps35129.4, Thaps35129.1, Thaps35129.6 |
| Expression Profile | Conditional Changes | Cluster Dendrogram | Discovered Potential cis-Regulatory Motifs |
|---|---|---|---|
Thaps_hclust_0269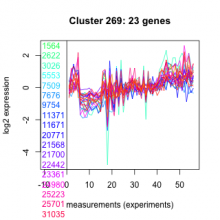 |
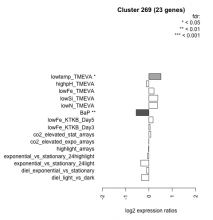 |
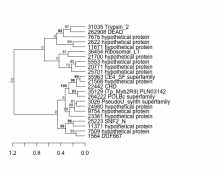 |
   |
| Normalized Mean Residue | Discovered Potential cis-Regulatory Motifs | |
|---|---|---|
|
Thaps_bicluster_0065 |
0.48 |
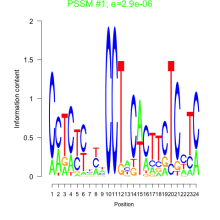 0.0000029 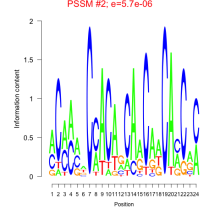 0.0000057 |
| KEGG description | KEGG Pathway |
|---|---|
| Not available | Not available |

Add comment