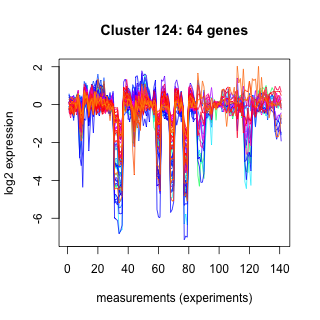
Phatr_hclust_0124 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0000226 | microtubule cytoskeleton organization and biogenesis | 0.0172156 | 1 | Phatr_hclust_0124 |
| GO:0006865 | amino acid transport | 0.0238392 | 1 | Phatr_hclust_0124 |
| GO:0006512 | ubiquitin cycle | 0.0420201 | 1 | Phatr_hclust_0124 |
| GO:0006304 | DNA modification | 0.0425004 | 1 | Phatr_hclust_0124 |
| GO:0006268 | DNA unwinding | 0.0425004 | 1 | Phatr_hclust_0124 |
| GO:0009073 | aromatic amino acid family biosynthesis | 0.0425004 | 1 | Phatr_hclust_0124 |
| GO:0006568 | tryptophan metabolism | 0.0507874 | 1 | Phatr_hclust_0124 |
| GO:0006821 | chloride transport | 0.0590046 | 1 | Phatr_hclust_0124 |
| GO:0009396 | folic acid and derivative biosynthesis | 0.0671528 | 1 | Phatr_hclust_0124 |
| GO:0006265 | DNA topological change | 0.0752324 | 1 | Phatr_hclust_0124 |
|
PHATRDRAFT_4413 : root border cell-specific protein (Pyrid_oxidase_2 superfamily) |
PHATRDRAFT_44333 : gamma-tubulin interacting protein (Spc97_Spc98) |
||
|
PHATRDRAFT_48186 : indole-3-glycerol-phosphate synthase (TIM_phosphate_binding superfamily) |
PHATRDRAFT_47881 : af225976_1 flavoredoxin (Flavin_Reduct) |
PHATRDRAFT_43515 : solute carrier family 29 (nucleoside transporters)memberisoform cra_b (Nucleoside_tran superfamily) |
PHATRDRAFT_15627 : ubiquitin-protein ligase (HECTc) |
|
PHATRDRAFT_50146 : amino acid transporter (Aa_trans superfamily) |
PHATRDRAFT_42424 : xenotropic and polytropic murine leukemia virus receptor xpr1 (EXS superfamily) |
||
|
PHATRDRAFT_43164 : short-chain dehydrogenase reductasesuperfamily protein (NADB_Rossmann superfamily) |
PHATRDRAFT_13261 : at4g39970 t5j17_140 (PLN02779) |
PHATRDRAFT_43807 : (DUF819 superfamily) |
|
|
PHATRDRAFT_46097 : chloride channel 7 (Voltage_gated_ClC superfamily) |
PHATRDRAFT_44868 : leucine aminopeptidase (PRK00913) |
PHATRDRAFT_18197 : atp-bindingsub-family c (cftr mrp)member 1 (ABCC_MRP_domain2) |
|
|
PHATRDRAFT_24119 : chloroplast light harvesting protein isoform 5 (Chloroa_b-bind) |
PHATRDRAFT_42606 : (SET) |
PHATRDRAFT_42123 : ubiquitin protein ligase (HECTc) |
|
|
PHATRDRAFT_18323 : inositol-phosphate phosphatase (IMPase) |
PHATRDRAFT_43280 : solute carrier familymember 1 (Aa_trans superfamily) |
PHATRDRAFT_43277 : chorismate mutase (PLN02344) |
PHATRDRAFT_32314 : short-chain dehydrogenase reductase sdr (NADB_Rossmann superfamily) |
|
PHATRDRAFT_23557 : cation-efflux transporter (CzcD) |
PHATRDRAFT_12732 : adenylate kinase cytosolic (adk) |
PHATRDRAFT_38927 : ribosomal protein l11 methyltransferase |
PHATRDRAFT_47117 : (Gly_transf_sug superfamily) |
|
PHATRDRAFT_45572 : ero1-like beta (ERO1 superfamily) |
PHATRDRAFT_46939 : protection of telomeres 1(pot1-like telomere end-binding protein) (RPA_2b-aaRSs_OBF_like superfamily) |
PHATRDRAFT_9115 : mgc116538 protein (AdoMet_MTases superfamily) |
|
|
PHATRDRAFT_33037 : (Methyltransf_28) |
PHATRDRAFT_18945 : at4g27720 t29a15_210 (MFS_1) |
PHATRDRAFT_15239 : otuubiquitin aldehyde binding 1 (Peptidase_C65) |
|
|
PHATRDRAFT_51271 : constans interacting protein 3 (FKBP_C) |
PHATRDRAFT_36077 : (Ten1_2 superfamily) |
PHATRDRAFT_42749 : component of oligomeric golgi complex 5 isoform 3 |
PHATRDRAFT_44738 : transmembrane emp24 domain-containing protein 9 precursor (EMP24_GP25L) |
|
PHATRDRAFT_12107 : dihydropterin pyrophosphokinase dihydropteroate synthase (DHPS) |
PHATRDRAFT_43317 : (DUF2088 superfamily) |
PHATRDRAFT_43435 : cog0457: fog: tpr repeat (COG4976) |
|
|
PHATRDRAFT_48187 : protein familywith a coiled coil-4 domain (Methyltransf_22) |
PHATRDRAFT_15125 : gdp-4-keto-6-deoxy-d-mannose epimerase-reductase (PLN02725) |
PHATRDRAFT_18335 : insulin-degrading enzyme (Ptr) |
|
|
PHATRDRAFT_33921 : (DUF4281) |
PHATRDRAFT_44905 : dolichyl glycosyltransferase (Alg6_Alg8 superfamily) |
PHATRDRAFT_43174 : gdp-fucose transporter (VRG4) |
PHATRDRAFT_44222 : enhanced disease susceptibility 5 (MATE_like superfamily) |
|
PHATRDRAFT_17086 : phosphoglycerate mutase 1 family (HP_PGM_like) |
PHATRDRAFT_9625 : presenilin family protein (Peptidase_A22B superfamily) |
PHATRDRAFT_13157 : small gtp-binding protein domain containing protein (Ras_like_GTPase superfamily) |
|
|
PHATRDRAFT_35643 : ornithine cyclodeaminase (PRK06199 superfamily) |
PHATRDRAFT_44480 : mgc82690 protein (MFS) |
PHATRDRAFT_8810 : topoisomeraseiii beta (TopA) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | 0.328 | 0.0070922 |
| Copper_SH | Copper_SH | 0.173 | 0.00748503 |
| Green_vs_Red_24h | Green_vs_Red_24h | -0.004 | 0.97885 |
| Silver_SH | Silver_SH | 0.123 | 0.045614 |
| Cadmium_1.2mg | Cadmium_1.2mg | -0.048 | 0.27512 |
| BlueLight_24h | BlueLight_24h | -0.085 | 0.725497 |
| GreenLight_0.5h | GreenLight_0.5h | -2.321 | 0.000458716 |
| RedLight_6h | RedLight_6h | 0.170 | 0.213346 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 0.980594 |
| GreenLight_24h | GreenLight_24h | -0.157 | 0.102899 |
| light_6hr | light_6hr | -0.046 | 0.778018 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | 0.022 | 0.894518 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.092 | 0.097 |
| Ammonia_SH | Ammonia_SH | -1.159 | 0.000344828 |
| Simazine_SH | Simazine_SH | -0.238 | 0.000423729 |
| Oil_SH | Oil_SH | 0.035 | 0.585337 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | 0.132 | 0.0811765 |
| Cadmium_SH | Cadmium_SH | -0.246 | 0.00217391 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | 0.067 | 0.514773 |
| highlight_0to6h | highlight_0to6h | -0.027 | 0.93125 |
| BlueLight_0.5h | BlueLight_0.5h | -3.104 | 0.000462963 |
| Dispersant_SH | Dispersant_SH | 0.000 | 0.999799 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | 0.191 | 0.00833333 |
| light_15.5hr | light_15.5hr | 0.385 | 0.00436893 |
| light_10.5hr | light_10.5hr | -0.020 | 0.909708 |
| Dispersed_oil_SH | Dispersed_oil_SH | 0.062 | 0.3956 |
| Si_free | Si_free | -0.053 | 0.512452 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | -3.030 | 0.000396825 |
| Mixture_SH | Mixture_SH | -1.255 | 0.000357143 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | -0.006 | 0.985113 |
| light_16hr_dark_30min | light_16hr_dark_30min | 0.467 | 0.000735294 |
| BlueLight_6h | BlueLight_6h | 0.164 | 0.094375 |
| highlight_12to48h | highlight_12to48h | 0.293 | 0.00634328 |
| RedLight_24h | RedLight_24h | -0.153 | 0.120924 |
| Dark_treated | Dark_treated | -2.262 | 0.000446429 |
| GreenLight_6h | GreenLight_6h | 0.256 | 0.0127219 |
| Cadmium_0.12mg | Cadmium_0.12mg | 0.023 | 0.376744 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | 0.072 | 0.0975248 |
| RedLight_0.5h | RedLight_0.5h | -1.756 | 0.000555556 |
| Green_vs_Red_6h | Green_vs_Red_6h | 0.086 | 0.547899 |
| Re-illuminated_24h | Re-illuminated_24h | 0.103 | 0.295608 |