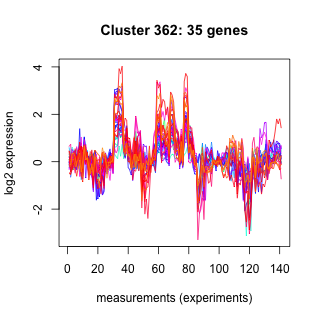
Phatr_hclust_0362 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006813 | potassium ion transport | 0.00148543 | 1 | Phatr_hclust_0362 |
| GO:0006811 | ion transport | 0.032161 | 1 | Phatr_hclust_0362 |
| GO:0016192 | vesicle-mediated transport | 0.0379055 | 1 | Phatr_hclust_0362 |
| GO:0008643 | carbohydrate transport | 0.0464638 | 1 | Phatr_hclust_0362 |
| GO:0006629 | lipid metabolism | 0.0521304 | 1 | Phatr_hclust_0362 |
| GO:0006812 | cation transport | 0.0772504 | 1 | Phatr_hclust_0362 |
| GO:0007242 | intracellular signaling cascade | 0.117765 | 1 | Phatr_hclust_0362 |
| GO:0016567 | protein ubiquitination | 0.193944 | 1 | Phatr_hclust_0362 |
| GO:0006810 | transport | 0.440472 | 1 | Phatr_hclust_0362 |
| GO:0006118 | electron transport | 0.503223 | 1 | Phatr_hclust_0362 |
|
PHATRDRAFT_2808 : protein disulfide isomerase associated 6 (Thioredoxin_like superfamily) |
PHATRDRAFT_45163 : (Glyco_tranf_2_4) |
PHATRDRAFT_36001 : ac027656_21cg10338 gene product from drosophila melanogaster gb (DUF647) |
|
|
PHATRDRAFT_49017 : telo2 protein (Telomere_reg-2 superfamily) |
|||
|
PHATRDRAFT_18469 : d-xylose proton-symporter (Sugar_tr) |
PHATRDRAFT_43150 : (RING) |
||
|
PHATRDRAFT_6292 : u5snrp brr2 sfii rna helicase (sec63 and the second part of the rna (HELICc) |
PHATRDRAFT_50123 : kef-type k+ transportnad-binding component (Ion_trans) |
||
|
PHATRDRAFT_37711 : (Lipase superfamily) |
PHATRDRAFT_41450 : heterogeneous nuclear ribonucleoprotein (RRM_SF superfamily) |
||
|
PHATRDRAFT_42542 : gaba-a receptor-associated membrane protein 3 (zf-DHHC superfamily) |
PHATRDRAFT_45401 : phoxdomain-containing protein (PX_SNX_like) |
PHATRDRAFT_21996 : nonsense-mediated mrna decay protein (NMD3) |
|
|
PHATRDRAFT_45223 : transmembrane protein 30a (CDC50) |
PHATRDRAFT_17683 : synaptobrevin-like protein (Synaptobrevin) |
PHATRDRAFT_36608 : nitroreductase (Nitro_FMN_reductase superfamily) |
PHATRDRAFT_44112 : enhanced disease susceptibility salicylic acid induction deficient (MATE_like superfamily) |
|
PHATRDRAFT_3676 : outward rectifying potassium channel kco (Ion_trans_2) |
PHATRDRAFT_49462 : diacylglycerol (LPLAT_MGAT-like) |
PHATRDRAFT_52444 : atcen2 calcium ion binding (PTZ00183) |
|
|
PHATRDRAFT_44111 : mopfamily transporter (MATE_like superfamily) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | 0.834 | 0.000505051 |
| Copper_SH | Copper_SH | -0.016 | 0.860105 |
| Green_vs_Red_24h | Green_vs_Red_24h | -0.377 | 0.00178571 |
| Silver_SH | Silver_SH | 0.083 | 0.300281 |
| Cadmium_1.2mg | Cadmium_1.2mg | 0.040 | 0.568439 |
| BlueLight_24h | BlueLight_24h | 0.070 | 0.802443 |
| GreenLight_0.5h | GreenLight_0.5h | 1.440 | 0.000458716 |
| RedLight_6h | RedLight_6h | 0.882 | 0.000694444 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 1 |
| GreenLight_24h | GreenLight_24h | 0.184 | 0.116547 |
| light_6hr | light_6hr | 0.415 | 0.0218935 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | 0.293 | 0.0667526 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.090 | 0.268373 |
| Ammonia_SH | Ammonia_SH | -0.797 | 0.000662252 |
| Simazine_SH | Simazine_SH | 0.334 | 0.00143885 |
| Oil_SH | Oil_SH | -0.201 | 0.0259259 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | 0.104 | 0.268803 |
| Cadmium_SH | Cadmium_SH | -0.064 | 0.508732 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | -0.490 | 0.00166667 |
| highlight_0to6h | highlight_0to6h | 0.081 | 0.658997 |
| BlueLight_0.5h | BlueLight_0.5h | 1.713 | 0.000462963 |
| Dispersant_SH | Dispersant_SH | 0.208 | 0.0251572 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | -0.155 | 0.112045 |
| light_15.5hr | light_15.5hr | -0.244 | 0.220492 |
| light_10.5hr | light_10.5hr | 0.340 | 0.0959906 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.257 | 0.00921053 |
| Si_free | Si_free | 0.030 | 0.776005 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | 1.915 | 0.000396825 |
| Mixture_SH | Mixture_SH | -1.114 | 0.000357143 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | -0.216 | 0.181579 |
| light_16hr_dark_30min | light_16hr_dark_30min | -0.290 | 0.131283 |
| BlueLight_6h | BlueLight_6h | 0.666 | 0.000581395 |
| highlight_12to48h | highlight_12to48h | -0.327 | 0.0260479 |
| RedLight_24h | RedLight_24h | 0.560 | 0.000943396 |
| Dark_treated | Dark_treated | 1.733 | 0.000446429 |
| GreenLight_6h | GreenLight_6h | 0.755 | 0.000609756 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.028 | 0.438889 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | -0.113 | 0.0666667 |
| RedLight_0.5h | RedLight_0.5h | 1.377 | 0.000555556 |
| Green_vs_Red_6h | Green_vs_Red_6h | -0.126 | 0.464155 |
| Re-illuminated_24h | Re-illuminated_24h | 0.284 | 0.00412844 |