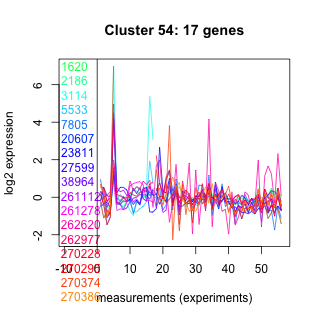
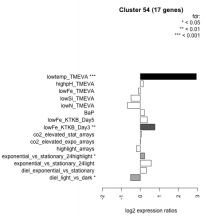
Thaps_hclust_0054 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009228 | thiamin biosynthetic process | 0.00165392 | 1 | Thaps_hclust_0054 |
| GO:0009073 | aromatic amino acid family biosynthesis | 0.0115273 | 1 | Thaps_hclust_0054 |
| GO:0045454 | cell redox homeostasis | 0.0147993 | 1 | Thaps_hclust_0054 |
| GO:0008299 | isoprenoid biosynthesis | 0.0180619 | 1 | Thaps_hclust_0054 |
| GO:0009765 | photosynthesis light harvesting | 0.0643185 | 1 | Thaps_hclust_0054 |
| GO:0006118 | electron transport | 0.0677231 | 1 | Thaps_hclust_0054 |
| GO:0006508 | proteolysis and peptidolysis | 0.38827 | 1 | Thaps_hclust_0054 |
|
1620 : S2P-M50 superfamily |
20607 : hypothetical protein |
261112 : Nudix_Hydrolase superfamily |
270228 : (Lhcx4) fucoxanthin chlorophyll a/c protein, LI818 clade |
|
2186 : ERG4_ERG24 superfamily |
23811 : ABC_ATPase superfamily |
261278 : GRX_GRXh_1_2_like |
270295 : hypothetical protein |
|
3114 : PRX_BCP |
27599 : hypothetical protein |
262620 : ANK |
270374 : hypothetical protein |
|
5533 : Chloroa_b-bind |
38964 : Chorismate_synthase |
262977 : NA |
270386 : hypothetical protein |
|
7805 : Aldo_ket_red |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | -0.509 | 0.016 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.767 | 0.00161 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.367 | 0.088 |
| BaP | BaP | 0.215 | 0.386 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.228 | 0.0437 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.053 | 0.856 |
| lowtemp_TMEVA | lowtemp_TMEVA | 2.950 | 0.000735 |
| highpH_TMEVA | highpH_TMEVA | 0.161 | 0.358 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.008 | 0.985 |
| lowFe_TMEVA | lowFe_TMEVA | -0.098 | 0.813 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.572 | 0.0843 |
| lowN_TMEVA | lowN_TMEVA | -0.675 | 0.0517 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.318 | 0.0972 |
| lowSi_TMEVA | lowSi_TMEVA | -0.487 | 0.372 |
| highlight_arrays | highlight_arrays | -0.202 | 0.244 |